CellChat example
Last updated: 2020-11-24
Checks: 7 0
Knit directory: interaction-tools/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20191213) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 0333fe3. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: .drake/
Ignored: data/COMUNET/
Ignored: data/CellChat/
Ignored: data/ICELLNET/
Ignored: data/NicheNet/
Ignored: data/cellphonedb/
Ignored: data/celltalker/
Ignored: output/14-CellChat.Rmd/
Ignored: output/15-talklr.Rmd/
Ignored: output/16-CiteFuse.Rmd/
Ignored: output/17-scTHI.Rmd/
Ignored: output/18-celltalker.Rmd/
Ignored: output/index.Rmd/
Ignored: renv/library/
Ignored: renv/python/
Ignored: renv/staging/
Unstaged changes:
Modified: _drake.R
Modified: output/11-CellPhoneDB.Rmd/count_network.txt
Modified: output/11-CellPhoneDB.Rmd/dotplot.png
Modified: output/11-CellPhoneDB.Rmd/heatmap_counts.png
Modified: output/11-CellPhoneDB.Rmd/heatmap_logcounts.png
Modified: output/11-CellPhoneDB.Rmd/interactions_count.txt
Modified: output/11-CellPhoneDB.Rmd/pvalues.txt
Modified: output/11-CellPhoneDB.Rmd/significant_means.txt
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/14-CellChat.Rmd) and HTML (docs/14-CellChat.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 9940762 | Luke Zappia | 2020-09-24 | Add talklr to drake |
| Rmd | b6db7b4 | Luke Zappia | 2020-07-30 | Add CellChat walkthrough |
| html | b6db7b4 | Luke Zappia | 2020-07-30 | Add CellChat walkthrough |
| Rmd | f017448 | Luke Zappia | 2020-07-29 | Setup CellChat |
| html | f017448 | Luke Zappia | 2020-07-29 | Setup CellChat |
# Setup document
source(here::here("code", "setup.R"))
# Function dependencies
invisible(drake::readd(download_link))Introduction
In this document we are going to run through the example analysis for the CellChat package and have a look at the output it produces. More information about CellChat can be found at https://github.com/sqjin/CellChat.
library("CellChat")
library("ggalluvial")Chunk time: 2.71 secs
1 Input
CellChat requires two user inputs: an expression matrix and either cell-type labels or a low-dimensional embedding. Here we use the label-based mode. The other inputs to CellChat are the ligand-receptor database and protein-protein interaction network included in the package.
load(fs::path(PATHS$CellChat_in, "data_wound_CellChat.rda"))Chunk time: 1.91 secs
1.1 Expression matrix
The expression matrix should be normalised with genes as rows and cells as columns. Here is a snippet of the example dataset:
expr <- data_wound$data
pander(as.matrix(expr[1:5, 1:5]))| AAACCTGAGATGTGGC | AAACCTGAGGTGTTAA | AAACCTGAGTATCTCG | |
|---|---|---|---|
| Xkr4 | 0 | 0 | 0 |
| Sox17 | 0 | 0 | 0 |
| Mrpl15 | 0 | 1.724 | 0 |
| Lypla1 | 0 | 0 | 0 |
| Tcea1 | 1.432 | 0 | 0 |
| AAACCTGCAAAGCAAT | AAACCTGCAGCTTCGG | |
|---|---|---|
| Xkr4 | 0 | 0 |
| Sox17 | 0 | 0 |
| Mrpl15 | 0 | 0 |
| Lypla1 | 0 | 0 |
| Tcea1 | 1.418 | 0 |
Chunk time: 0.14 secs
The full matrix has 17090 rows (genes) and 21557 columns (cells).
1.2 Labels
The second input is a data.frame with cell type labels for each cell.
labels <- data.frame(
group = data_wound$labels,
row.names = names(data_wound$labels)
)
group_size <- as.numeric(table(labels$group))
skim(labels)| Name | labels |
| Number of rows | 21557 |
| Number of columns | 1 |
| _______________________ | |
| Column type frequency: | |
| factor | 1 |
| ________________________ | |
| Group variables | None |
Variable type: factor
| skim_variable | n_missing | complete_rate | ordered | n_unique | top_counts |
|---|---|---|---|---|---|
| group | 0 | 1 | FALSE | 25 | FIB: 5073, FIB: 2817, FIB: 1957, FIB: 1323 |
Chunk time: 0.06 secs
1.3 Database
The database is available as a list. The tutorial suggests using only the interactions annotated with “Secreted Signalling”.
CellChatDB <- CellChatDB.mouse
CellChatDB <- subsetDB(CellChatDB, search = "Secreted Signaling")Chunk time: 0.15 secs
The database list contains 4 items: interaction, complex, cofactor and geneInfo
1.3.1 Interaction
The first item in the list is a data.frame with information about interactions.
skim(CellChatDB$interaction)| Name | CellChatDB$interaction |
| Number of rows | 1211 |
| Number of columns | 11 |
| _______________________ | |
| Column type frequency: | |
| character | 11 |
| ________________________ | |
| Group variables | None |
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| interaction_name | 0 | 1 | 6 | 21 | 0 | 1211 | 0 |
| pathway_name | 0 | 1 | 2 | 12 | 0 | 141 | 0 |
| ligand | 0 | 1 | 2 | 12 | 0 | 361 | 0 |
| receptor | 0 | 1 | 3 | 18 | 0 | 363 | 0 |
| agonist | 0 | 1 | 0 | 15 | 868 | 5 | 0 |
| antagonist | 0 | 1 | 0 | 18 | 755 | 11 | 0 |
| co_A_receptor | 0 | 1 | 0 | 25 | 829 | 9 | 0 |
| co_I_receptor | 0 | 1 | 0 | 27 | 724 | 10 | 0 |
| evidence | 0 | 1 | 12 | 45 | 0 | 376 | 0 |
| annotation | 0 | 1 | 18 | 18 | 0 | 1 | 0 |
| interaction_name_2 | 0 | 1 | 9 | 26 | 0 | 1211 | 0 |
Chunk time: 0.08 secs
1.3.2 Complex
The second item contain information about complexes.
skim(CellChatDB$complex)| Name | CellChatDB$complex |
| Number of rows | 155 |
| Number of columns | 4 |
| _______________________ | |
| Column type frequency: | |
| character | 4 |
| ________________________ | |
| Group variables | None |
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| subunit_1 | 0 | 1 | 3 | 7 | 0 | 96 | 0 |
| subunit_2 | 0 | 1 | 3 | 7 | 0 | 70 | 0 |
| subunit_3 | 0 | 1 | 0 | 6 | 142 | 10 | 0 |
| subunit_4 | 0 | 1 | 0 | 3 | 154 | 2 | 0 |
Chunk time: 0.05 secs
1.3.3 Cofactor
The next item contains information about cofactors.
skim(CellChatDB$cofactor)| Name | CellChatDB$cofactor |
| Number of rows | 31 |
| Number of columns | 16 |
| _______________________ | |
| Column type frequency: | |
| character | 16 |
| ________________________ | |
| Group variables | None |
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| cofactor1 | 0 | 1 | 2 | 6 | 0 | 25 | 0 |
| cofactor2 | 0 | 1 | 0 | 6 | 19 | 12 | 0 |
| cofactor3 | 0 | 1 | 0 | 6 | 22 | 9 | 0 |
| cofactor4 | 0 | 1 | 0 | 8 | 24 | 7 | 0 |
| cofactor5 | 0 | 1 | 0 | 6 | 25 | 6 | 0 |
| cofactor6 | 0 | 1 | 0 | 6 | 26 | 5 | 0 |
| cofactor7 | 0 | 1 | 0 | 6 | 28 | 4 | 0 |
| cofactor8 | 0 | 1 | 0 | 6 | 28 | 4 | 0 |
| cofactor9 | 0 | 1 | 0 | 5 | 29 | 3 | 0 |
| cofactor10 | 0 | 1 | 0 | 7 | 29 | 3 | 0 |
| cofactor11 | 0 | 1 | 0 | 5 | 30 | 2 | 0 |
| cofactor12 | 0 | 1 | 0 | 5 | 30 | 2 | 0 |
| cofactor13 | 0 | 1 | 0 | 6 | 30 | 2 | 0 |
| cofactor14 | 0 | 1 | 0 | 6 | 30 | 2 | 0 |
| cofactor15 | 0 | 1 | 0 | 7 | 30 | 2 | 0 |
| cofactor16 | 0 | 1 | 0 | 4 | 30 | 2 | 0 |
Chunk time: 0.09 secs
1.3.4 Gene info
The final item in the database is gene annotation information.
skim(CellChatDB$geneInfo)| Name | CellChatDB$geneInfo |
| Number of rows | 45544 |
| Number of columns | 6 |
| _______________________ | |
| Column type frequency: | |
| character | 6 |
| ________________________ | |
| Group variables | None |
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| Symbol | 0 | 1 | 1 | 16 | 0 | 45544 | 0 |
| Name | 0 | 1 | 5 | 139 | 0 | 45528 | 0 |
| EntrezGene.ID | 0 | 1 | 4 | 9 | 0 | 45536 | 1 |
| Ensembl.Gene.ID | 0 | 1 | 0 | 18 | 5 | 36640 | 0 |
| HomoloGene.ID | 0 | 1 | 0 | 6 | 4 | 18941 | 0 |
| HGNC.ID | 0 | 1 | 0 | 85 | 2 | 17932 | 0 |
Chunk time: 0.25 secs
1.4 Network
The network is a binary matrix showing interactions between proteins.
pander(as.matrix(PPI.mouse[1:5, 1:5]))| Cdh1 | Bcl6b | Pparg | Raf1 | Kat2b | |
|---|---|---|---|---|---|
| Cdh1 | 0 | 0 | 0 | 0 | 0 |
| Bcl6b | 0 | 0 | 0 | 0 | 0 |
| Pparg | 0 | 0 | 0 | 0 | 0 |
| Raf1 | 0 | 0 | 0 | 0 | 0 |
| Kat2b | 0 | 0 | 0 | 0 | 0 |
Chunk time: 0.01 secs
The full matrix has 557 rows and 557 columns.
2 Create CellChat object
CellChat uses a custom object which we can create from the expression matrix and labels. We also store the database here.
cellchat <- createCellChat(expr)
cellchat <- addMeta(cellchat, meta = labels, meta.name = "labels")
cellchat <- setIdent(cellchat, ident.use = "labels")
cellchat@DB <- CellChatDB
cellchatAn object of class CellChat
17090 genes.
21557 cells.Chunk time: 0.01 secs
3 Pre-processing
3.1 Subset signalling genes
First we subset the object to select only the signalling genes that are listed in the database.
cellchat <- subsetData(cellchat)
cellchatAn object of class CellChat
17090 genes.
21557 cells.Chunk time: 0.19 secs
3.2 Identify over-expression
The next step is to identify over-expressed genes and interactions. Over-expressed genes are detected using a one-sided Wilcoxon test. Interactions are selected if either a ligand or receptor is over-expressed.
cellchat <- identifyOverExpressedGenes(cellchat)
cellchat <- identifyOverExpressedInteractions(cellchat)Chunk time: 4.16 mins
This step has identified 539 genes and 666 interactions.
3.3 Project data
The gene expression data is then projected onto a protein-protein interaction network.
cellchat <- projectData(cellchat, PPI.mouse)Chunk time: 1.63 secs
4 Communication network inference
4.1 Compute communication probability
At this stage we can now compute the communication probability and infer the communication network. The results are stored in the net slot of the CellChat object which contains three-dimensional arrays of probabilities and p-values for interactions between each pair of cell types for each interaction pair. Here are snippets of these arrays for the first interaction pair.
cellchat <- computeCommunProb(cellchat)
pander(cellchat@net$prob[1:5, 1:5, 1])| FIB-A | FIB-B | FIB-C | FIB-D | FIB-E | |
|---|---|---|---|---|---|
| FIB-A | 0 | 0 | 1.186e-07 | 1.152e-06 | 5.496e-07 |
| FIB-B | 0 | 0 | 0 | 0 | 0 |
| FIB-C | 0 | 0 | 8.06e-08 | 7.834e-07 | 3.737e-07 |
| FIB-D | 0 | 0 | 7.92e-08 | 7.707e-07 | 3.676e-07 |
| FIB-E | 0 | 0 | 5.006e-08 | 4.868e-07 | 2.322e-07 |
pander(cellchat@net$pval[1:5, 1:5, 1])| FIB-A | FIB-B | FIB-C | FIB-D | FIB-E | |
|---|---|---|---|---|---|
| FIB-A | 1 | 1 | 0 | 0 | 0 |
| FIB-B | 1 | 1 | 1 | 1 | 1 |
| FIB-C | 1 | 1 | 0 | 0 | 0 |
| FIB-D | 1 | 1 | 0.09 | 0 | 0 |
| FIB-E | 1 | 1 | 0.22 | 0 | 0 |
Chunk time: 4.56 mins
4.2 Infer pathway signalling
We also compute the communication probabilities at the pathway level by summarising all ligand-receptor interactions associated with each pathway. The output is similar but now instead of interaction pairs we have pathways.
cellchat <- computeCommunProbPathway(cellchat)
pander(cellchat@netP$prob[1:5, 1:5, 1])| FIB-A | FIB-B | FIB-C | FIB-D | FIB-E | |
|---|---|---|---|---|---|
| FIB-A | 0 | 0 | 1.186e-07 | 1.152e-06 | 5.496e-07 |
| FIB-B | 0 | 0 | 0 | 0 | 0 |
| FIB-C | 0 | 0 | 8.06e-08 | 7.834e-07 | 3.737e-07 |
| FIB-D | 0 | 0 | 8.283e-08 | 1.577e-06 | 7.521e-07 |
| FIB-E | 0 | 0 | 5.867e-08 | 1.057e-06 | 5.043e-07 |
Chunk time: 1.09 secs
4.3 Aggregate network
We can get an aggregated communication network by counting the number of links between cell types or summarising the communication probabilities. These results are stored as two-dimensional matrices.
cellchat <- aggregateNet(cellchat)
pander(cellchat@net$count[1:5, 1:5])| FIB-A | FIB-B | FIB-C | FIB-D | FIB-E | |
|---|---|---|---|---|---|
| FIB-A | 5 | 3 | 6 | 8 | 8 |
| FIB-B | 4 | 1 | 1 | 2 | 4 |
| FIB-C | 5 | 2 | 4 | 6 | 8 |
| FIB-D | 12 | 9 | 15 | 19 | 21 |
| FIB-E | 15 | 13 | 20 | 25 | 29 |
pander(cellchat@net$sum[1:5, 1:5])| FIB-A | FIB-B | FIB-C | FIB-D | FIB-E | |
|---|---|---|---|---|---|
| FIB-A | 4.806e-06 | 7.669e-07 | 1.322e-06 | 4.156e-06 | 2.752e-06 |
| FIB-B | 1.435e-05 | 3.489e-06 | 1.862e-06 | 2.748e-06 | 2.677e-06 |
| FIB-C | 4.64e-06 | 2.509e-06 | 1.434e-06 | 3.339e-06 | 2.769e-06 |
| FIB-D | 5.496e-05 | 0.0001328 | 7.748e-05 | 0.0001942 | 0.0001317 |
| FIB-E | 2.67e-05 | 7.208e-05 | 4.105e-05 | 0.0001038 | 7.044e-05 |
Chunk time: 0.04 secs
5 Network analysis
CellChat can perform analysis on the communication network to better understand the roles of each cell type. Here we perform that analysis using the pathway scores. The output is a list with a set of metrics for cell type for each pathway.
cellchat <- netAnalysis_signalingRole(cellchat, slot.name = "netP")Chunk time: 1.81 secs
The network metrics are: outdeg, indeg, hub, authority, eigen, page_rank, betweenness, flowbet and info
6 Global patterns
It may also be interesting to identify common communication patterns. We can do this for both outgoing and incoming patterns.
cellchat <- identifyCommunicationPatterns(cellchat, pattern = "outgoing", k = 5)
cellchat <- identifyCommunicationPatterns(cellchat, pattern = "incoming", k = 5)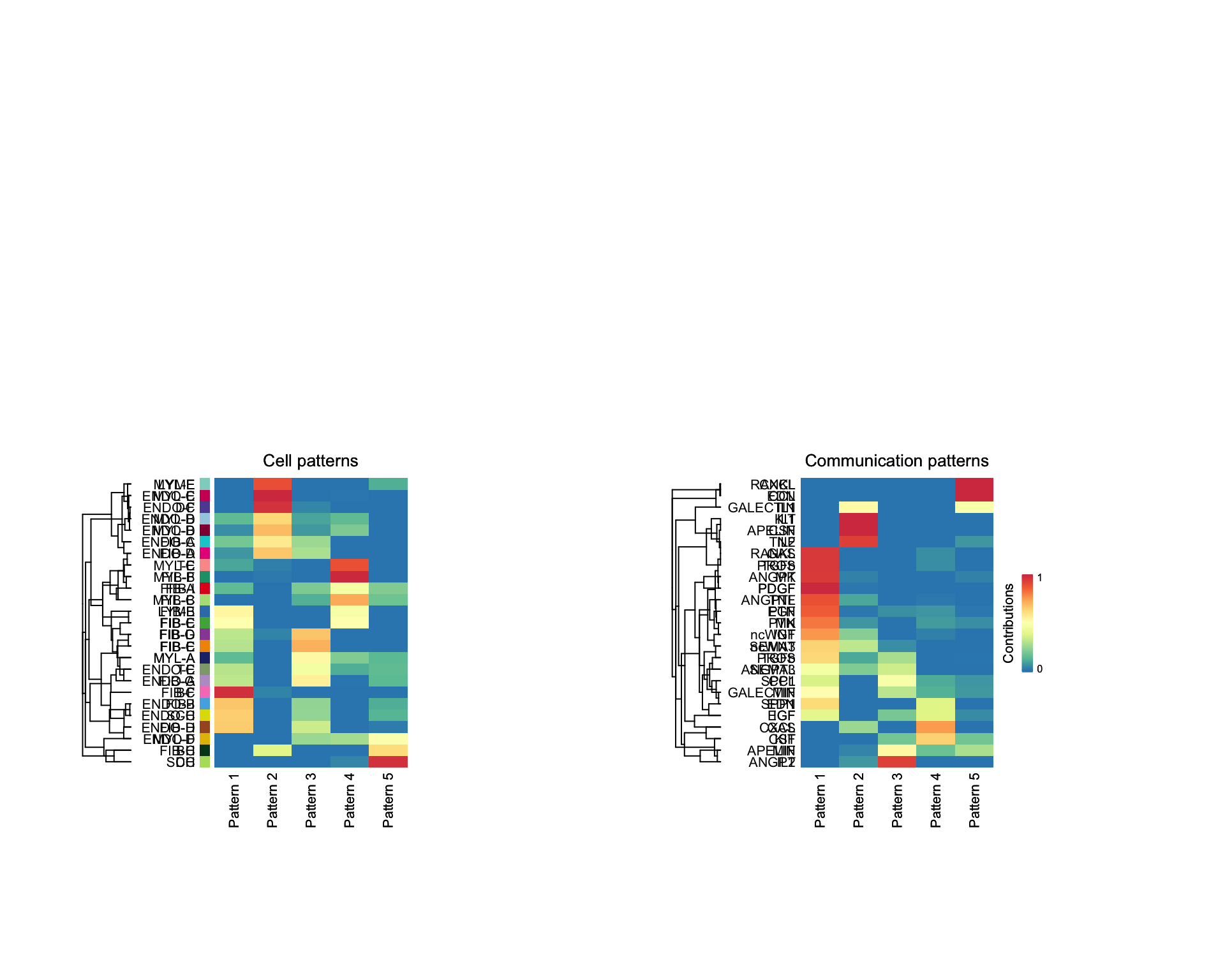
| Version | Author | Date |
|---|---|---|
| b6db7b4 | Luke Zappia | 2020-07-30 |
Chunk time: 6.79 secs
7 Manifold learning and classification
CellChat can identify groups of signalling pathways by embedding and clustering them.
7.1 Functional similarity
Functional similarity indicates that major senders and receivers are similar and can be interpreted as the two pathways or interaction pairs exhibiting similar and/or redundant roles.
The output is a similarity matrix, UMAP embedding and cluster assignments from \(k\)-means.
cellchat <- computeNetSimilarity(cellchat, type = "functional", thresh = 0.25)
cellchat <- netEmbedding(cellchat, type = "functional")
cellchat <- netClustering(cellchat, type = "functional", k = 4)
pander(cellchat@netP$similarity$functional$matrix[1:5,1:5])| TGFb | ncWNT | EGF | PDGF | IGF | |
|---|---|---|---|---|---|
| TGFb | 1 | 0 | 0.008282 | 0.03183 | 0.009311 |
| ncWNT | 0 | 1 | 0 | 0 | 0 |
| EGF | 0.008282 | 0 | 1 | 0.08182 | 0 |
| PDGF | 0.03183 | 0 | 0.08182 | 1 | 0 |
| IGF | 0.009311 | 0 | 0 | 0 | 1 |
pander(cellchat@netP$similarity$functional$dr[1:5, ])| UMAP1 | UMAP2 | |
|---|---|---|
| TGFb | -2.434 | 5.187 |
| ncWNT | -2.311 | 2.232 |
| EGF | -3.18 | 2.811 |
| PDGF | -2.357 | 3.149 |
| IGF | -5.963 | -2.334 |
cellchat@netP$similarity$functional$group TGFb ncWNT EGF PDGF IGF APELIN CCL CXCL
3 1 1 1 2 4 3 4
MIF IL1 CSF TNF SPP1 ANGPTL MK PTN
3 4 3 3 2 2 2 2
KIT SEMA3 GAS GALECTIN PROS
4 2 1 3 1 Chunk time: 10.23 secs
7.2 Structural similarity
Structural similarity is used to compare the signalling network structure, without considering the similarity of senders and receivers.
cellchat <- computeNetSimilarity(cellchat, type = "structural", thresh = 0.25)
cellchat <- netEmbedding(cellchat, type = "structural")
cellchat <- netClustering(cellchat, type = "structural", k = 4)
pander(cellchat@netP$similarity$structural$matrix[1:5,1:5])| TGFb | ncWNT | EGF | PDGF | IGF | |
|---|---|---|---|---|---|
| TGFb | 1 | 0 | 0 | 0 | 0 |
| ncWNT | 0 | 1 | 0.3642 | 0.5772 | 0.3458 |
| EGF | 0 | 0.3642 | 1 | 0.5978 | 0.5932 |
| PDGF | 0 | 0.5772 | 0.5978 | 1 | 0.6122 |
| IGF | 0 | 0.3458 | 0.5932 | 0.6122 | 1 |
pander(cellchat@netP$similarity$structural$dr[1:5, ])| UMAP1 | UMAP2 | |
|---|---|---|
| TGFb | -14.25 | -2.163 |
| ncWNT | 21.32 | -6.326 |
| EGF | 21.53 | -5.578 |
| PDGF | 21.78 | -5.973 |
| IGF | 20.43 | -5.638 |
cellchat@netP$similarity$structural$group TGFb ncWNT EGF PDGF IGF APELIN CCL CXCL
2 1 1 1 1 3 4 3
MIF IL2 IL1 CSF TNF RANKL SPP1 ANGPTL
2 3 4 1 1 3 4 2
ANGPT MK PTN EDN KIT SEMA3 GAS GALECTIN
3 2 2 3 3 1 3 4
PROS
3 Chunk time: 9.98 secs
8 Visualisation
8.1 Signalling pathways
8.1.1 Hierarchy plot
Here solid circles are sources and open circles are targets. Circle sizes are the number of cells in each group and lines are coloured according to the source with thicker lines indicating stronger signals.
netVisual_aggregate(cellchat, "TGFb", vertex.receiver = 1:9, # groups to show
vertex.size = group_size)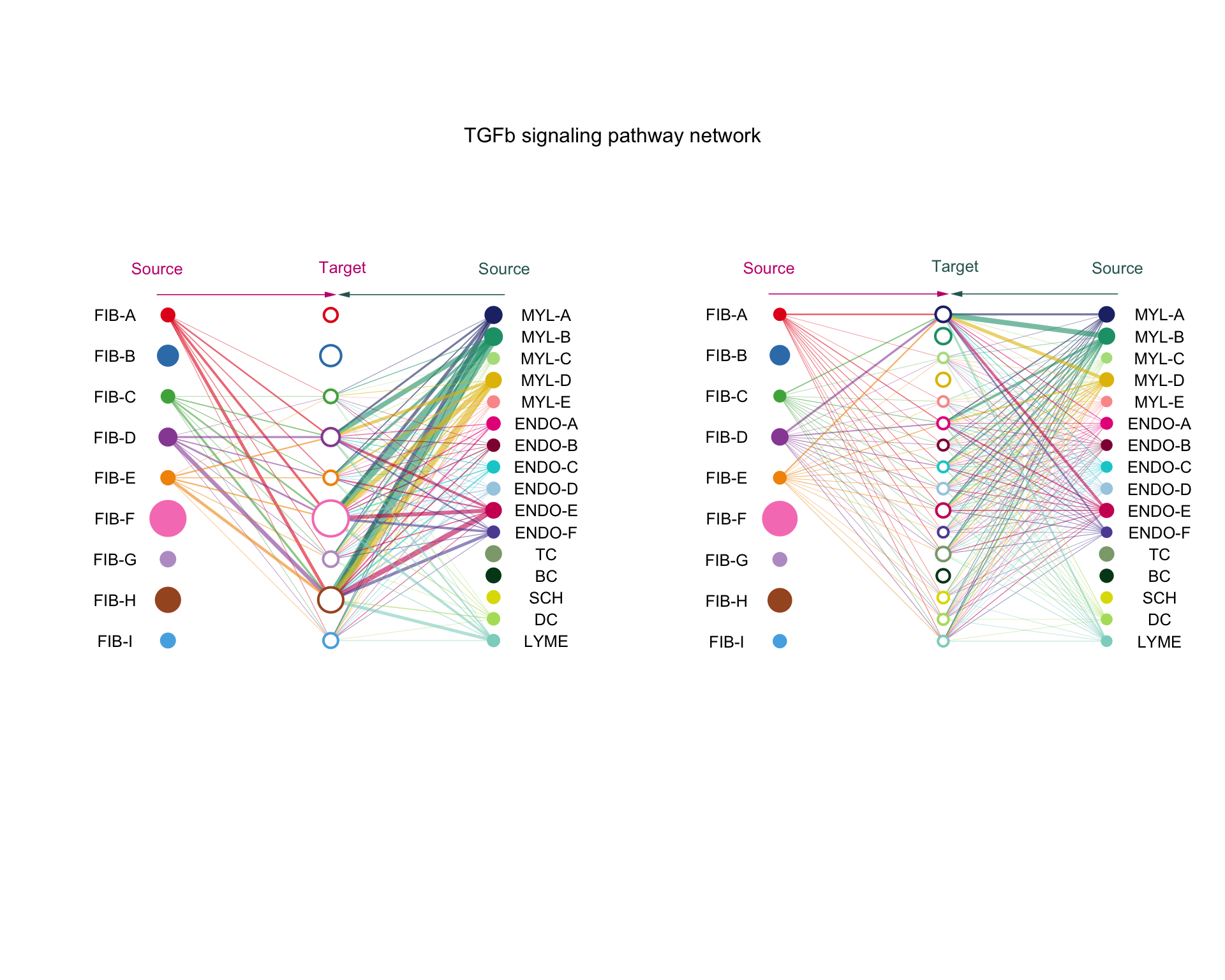
| Version | Author | Date |
|---|---|---|
| b6db7b4 | Luke Zappia | 2020-07-30 |
Chunk time: 0.36 secs
8.1.2 Circle plot
We can show a similar thing as a circle plot.
netVisual_aggregate(cellchat, "ncWNT", layout = "circle",
vertex.size = group_size)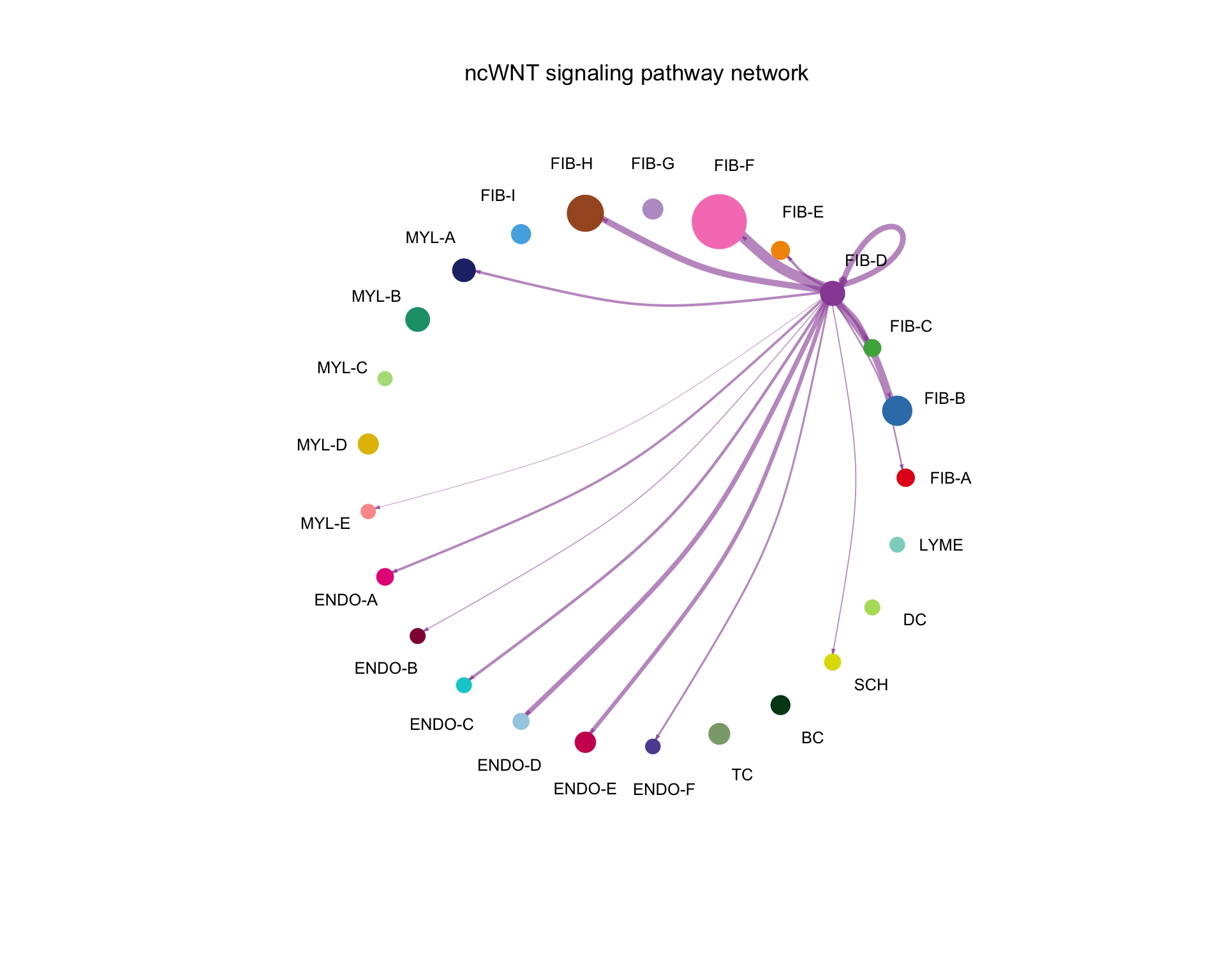
| Version | Author | Date |
|---|---|---|
| b6db7b4 | Luke Zappia | 2020-07-30 |
Chunk time: 0.18 secs
8.2 Pathway contribution
Here we show the contribution of each interaction pair to a signalling pathway.
netAnalysis_contribution(cellchat, "TGFb")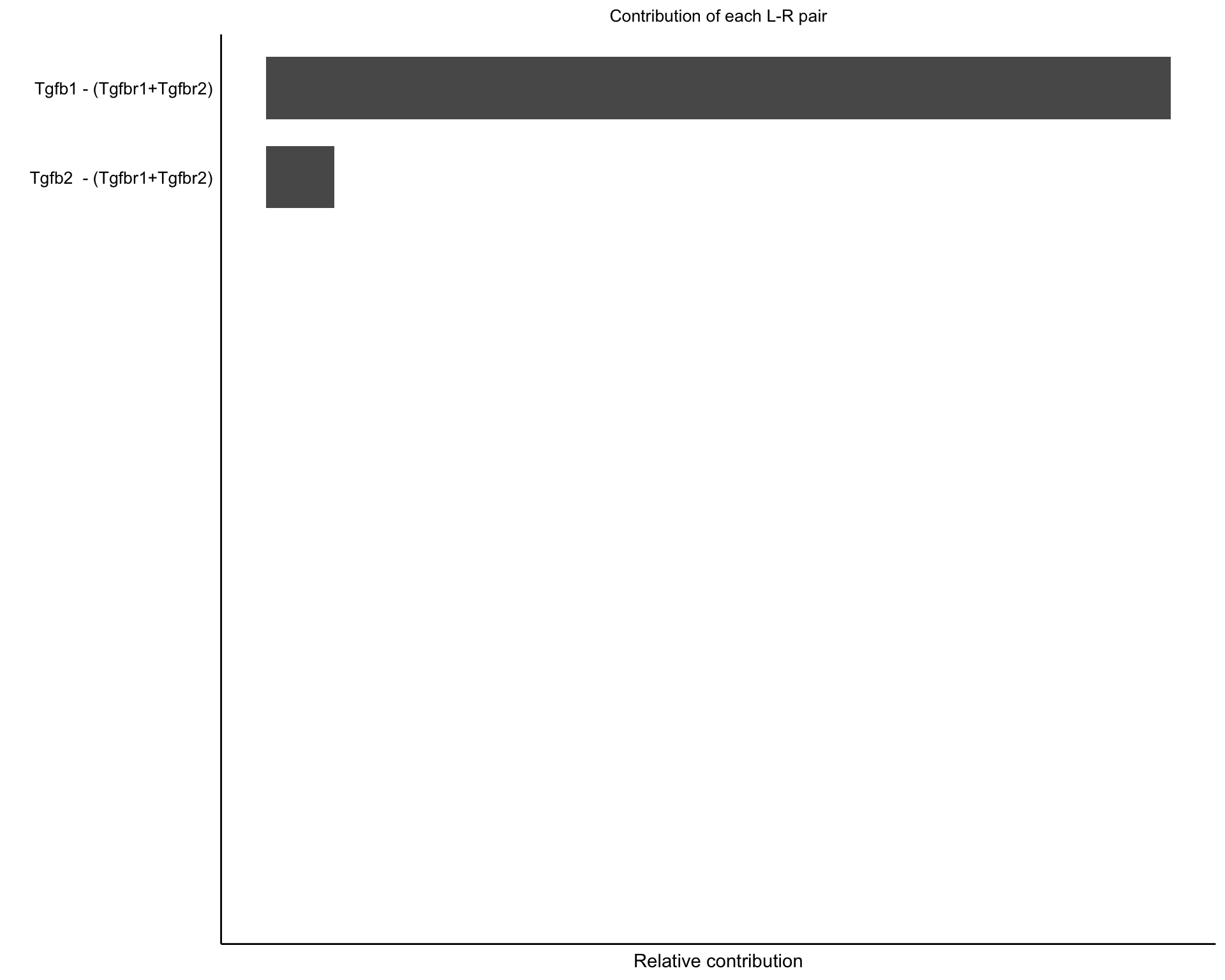
| Version | Author | Date |
|---|---|---|
| b6db7b4 | Luke Zappia | 2020-07-30 |
Chunk time: 0.3 secs
8.3 Signalling roles
For can look at the expected role for each cell type for a pathway based on the network analysis scores.
netVisual_signalingRole(cellchat, "TGFb", width = 16, height = 4,
font.size = 10)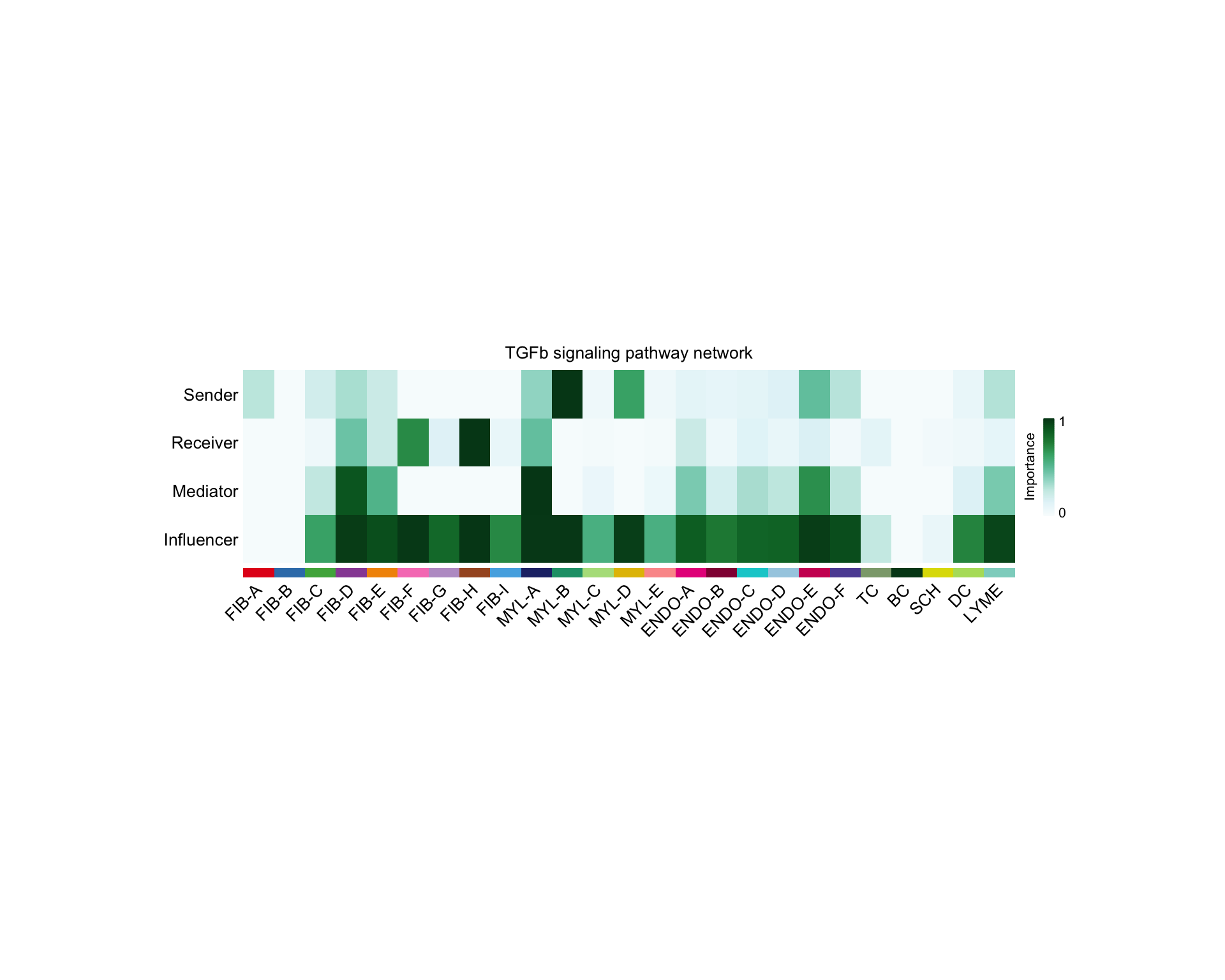
| Version | Author | Date |
|---|---|---|
| b6db7b4 | Luke Zappia | 2020-07-30 |
Chunk time: 0.38 secs
8.4 Communication patterns
8.4.1 River plot
An alternative way to visualise the communication patterns is using a river plot.
netAnalysis_river(cellchat, pattern = "outgoing")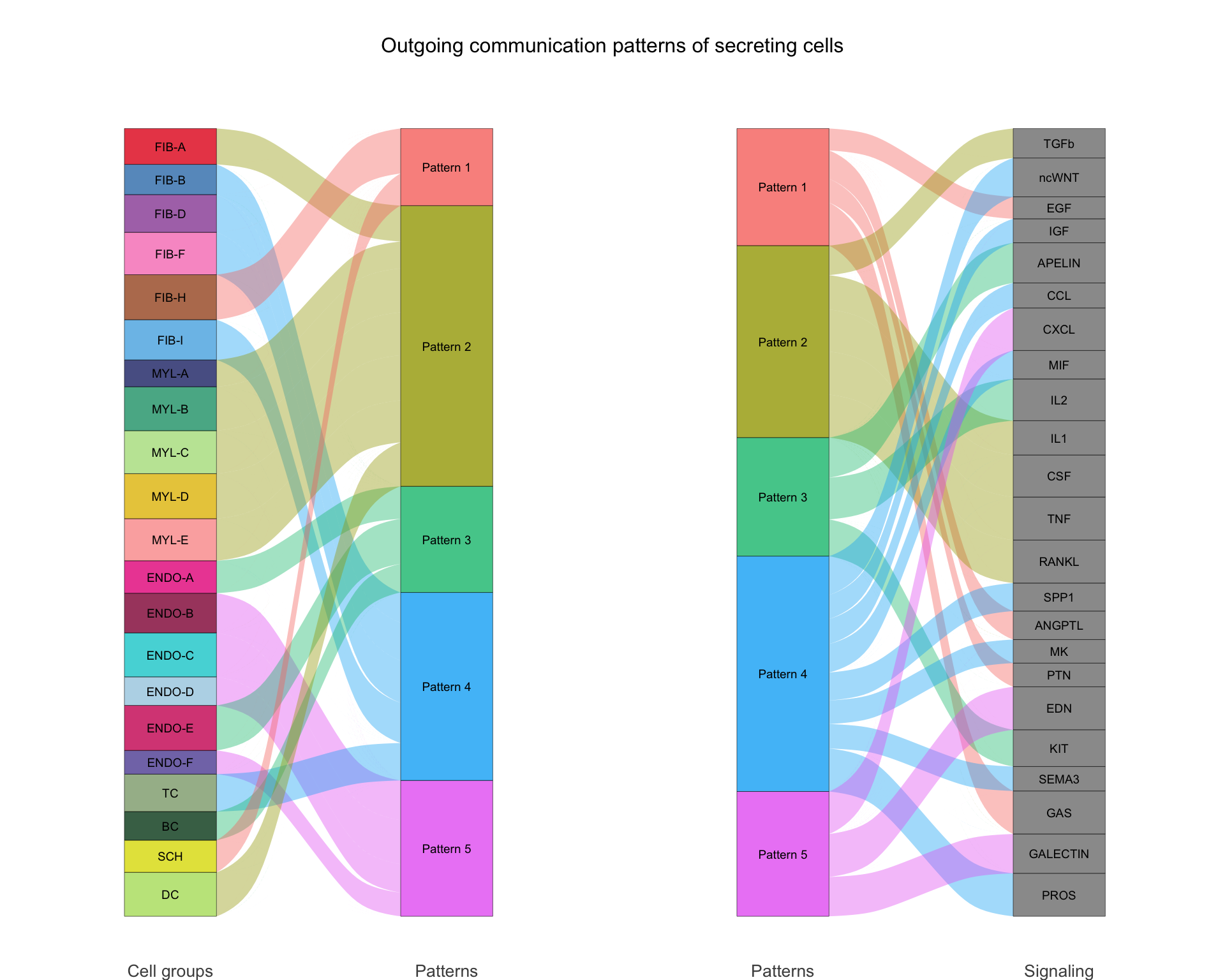
| Version | Author | Date |
|---|---|---|
| b6db7b4 | Luke Zappia | 2020-07-30 |
Chunk time: 1.26 secs
8.4.2 Dot plot
Another alternative is a dot plot.
netAnalysis_dot(cellchat, pattern = "outgoing")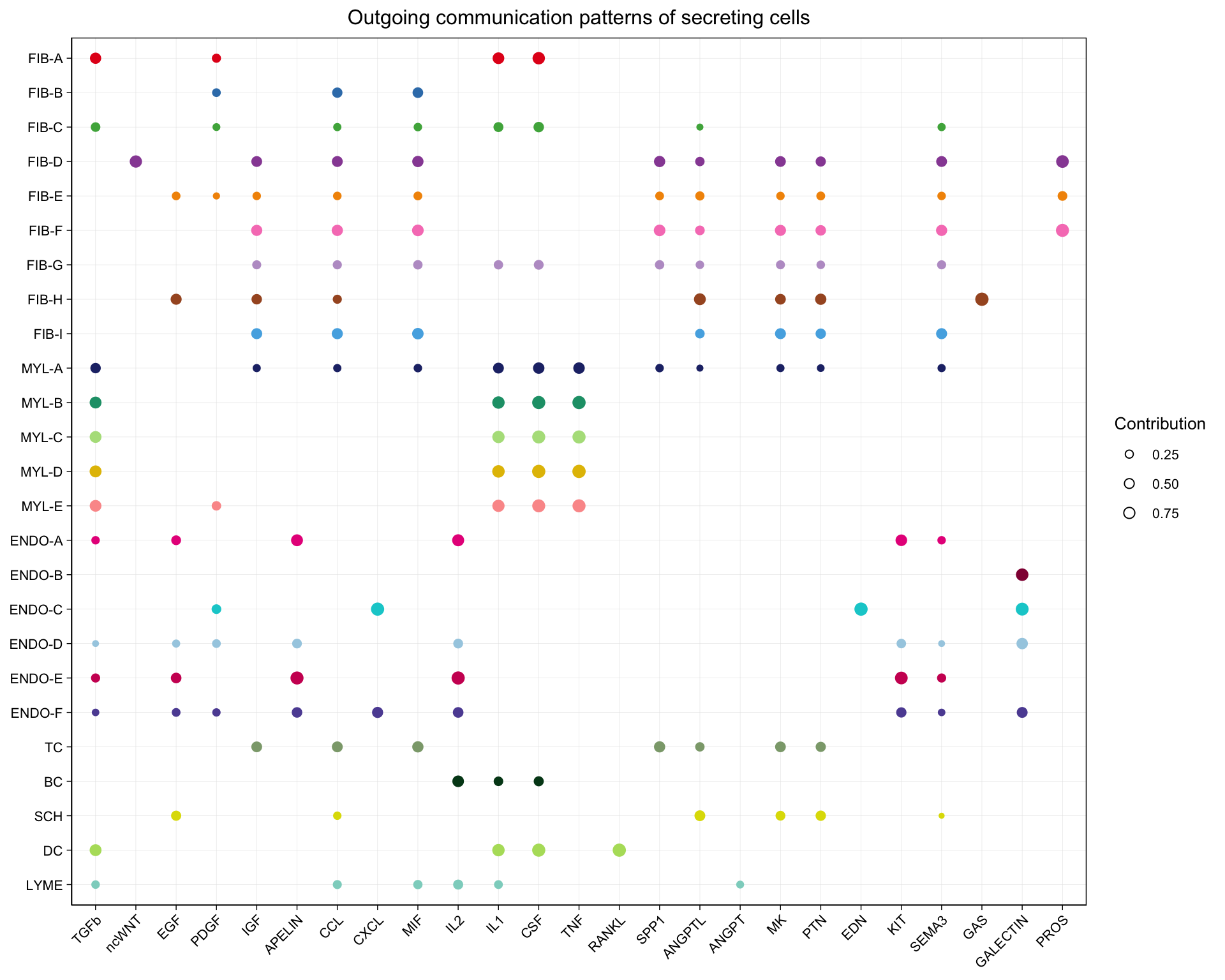
| Version | Author | Date |
|---|---|---|
| b6db7b4 | Luke Zappia | 2020-07-30 |
Chunk time: 0.56 secs
8.5 Embedding
UMAP embedding of pathway similarity.
netVisual_embedding(cellchat, type = "functional", pathway.remove.show = FALSE,
label.size = 3.5)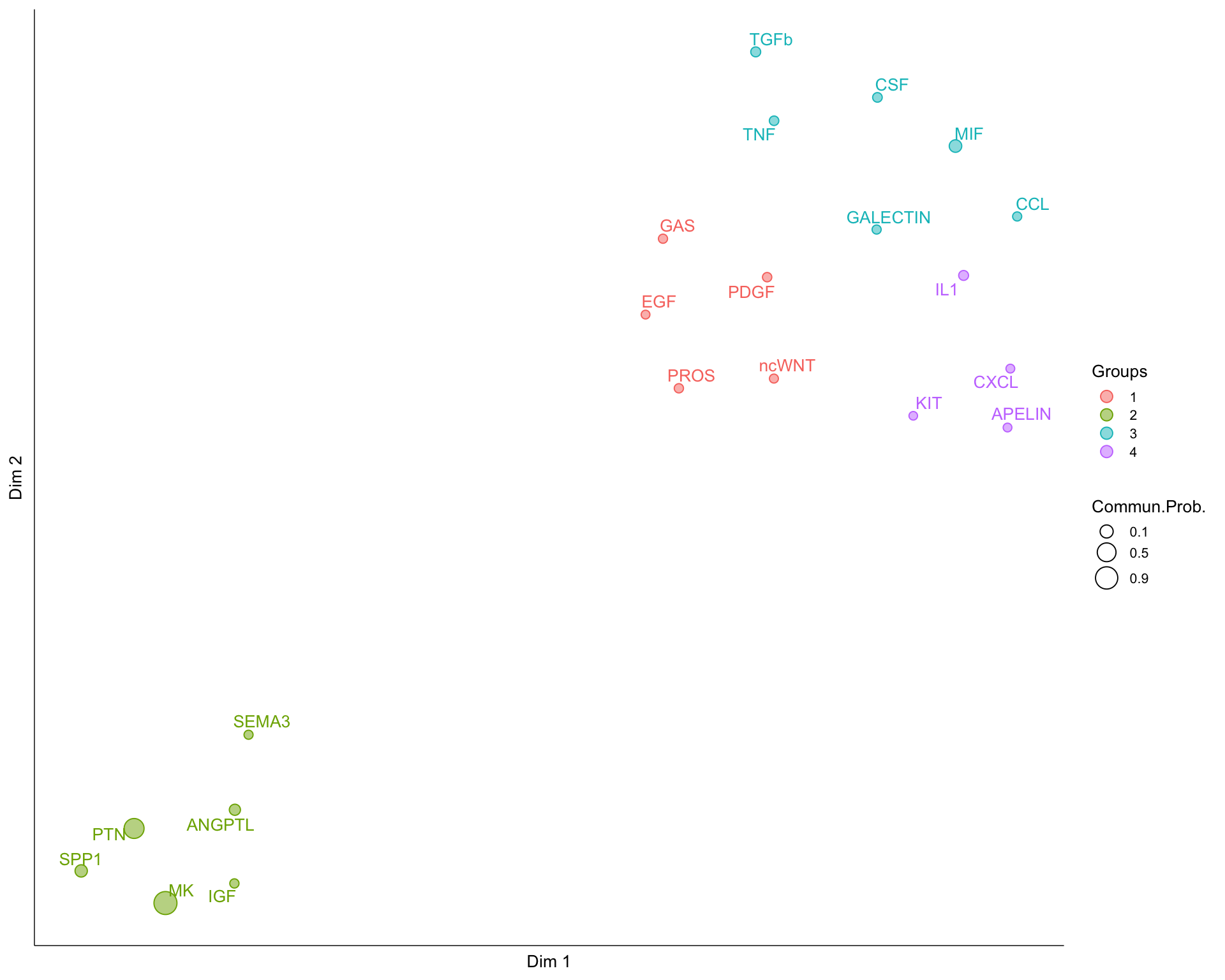
| Version | Author | Date |
|---|---|---|
| 9940762 | Luke Zappia | 2020-09-24 |
| b6db7b4 | Luke Zappia | 2020-07-30 |
Chunk time: 0.57 secs
8.5.1 Zoom in
netVisual_embeddingZoomIn(cellchat, type = "functional", nCol = 2)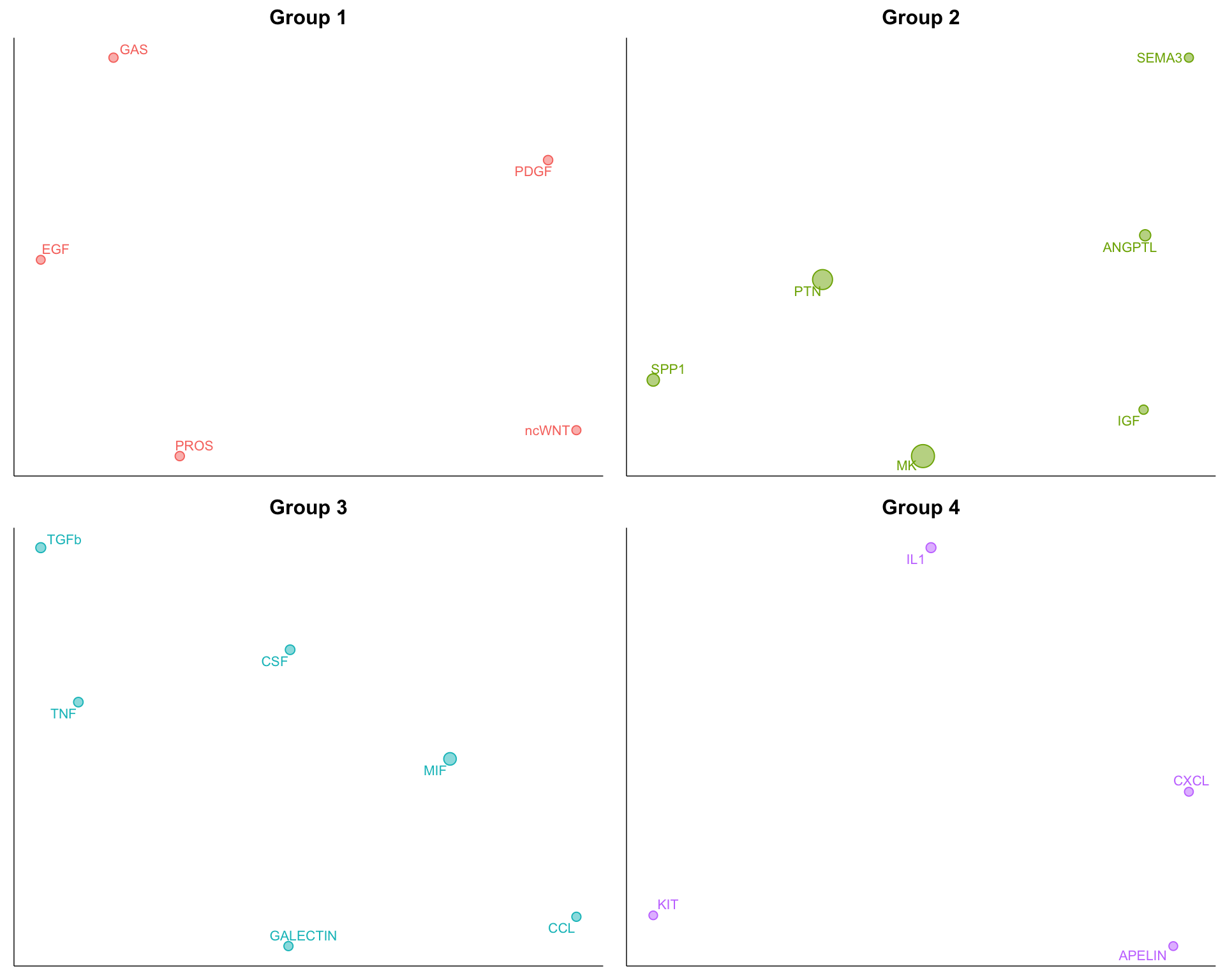
| Version | Author | Date |
|---|---|---|
| 9940762 | Luke Zappia | 2020-09-24 |
| b6db7b4 | Luke Zappia | 2020-07-30 |
Chunk time: 0.59 secs
Summary
Parameters
This table describes parameters used and set in this document.
params <- list(
)
params <- toJSON(params, pretty = TRUE)
kable(fromJSON(params))Chunk time: 0.01 secs
Output files
This table describes the output files produced by this document. Right click and Save Link As… to download the results.
kable(data.frame(
File = c(
download_link("parameters.json", OUT_DIR)
),
Description = c(
"Parameters set and used in this analysis"
)
))| File | Description |
|---|---|
| parameters.json | Parameters set and used in this analysis |
Chunk time: 0.01 secs
Session information
sessioninfo::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.0.0 (2020-04-24)
os macOS Catalina 10.15.7
system x86_64, darwin17.0
ui X11
language (EN)
collate en_US.UTF-8
ctype en_US.UTF-8
tz Europe/Berlin
date 2020-11-24
─ Packages ───────────────────────────────────────────────────────────────────
! package * version date lib
P assertthat 0.2.1 2019-03-21 [?]
P backports 1.1.6 2020-04-05 [?]
P base64enc 0.1-3 2015-07-28 [?]
P base64url 1.4 2018-05-14 [?]
bibtex 0.4.2.2 2020-01-02 [1]
Biobase * 2.48.0 2020-04-27 [1]
BiocGenerics * 0.34.0 2020-04-27 [1]
P broom 0.5.6 2020-04-20 [?]
P CellChat * 0.0.1 2020-07-29 [?]
P cellranger 1.1.0 2016-07-27 [?]
P circlize 0.4.9 2020-04-30 [?]
P cli 2.0.2 2020-02-28 [?]
clue 0.3-57 2019-02-25 [1]
P cluster 2.1.0 2019-06-19 [?]
coda 0.19-3 2019-07-05 [1]
P codetools 0.2-18 2020-11-04 [?]
P colorspace 1.4-1 2019-03-18 [?]
P ComplexHeatmap 2.5.4 2020-07-29 [?]
P conflicted * 1.0.4 2019-06-21 [?]
P cowplot 1.0.0 2019-07-11 [?]
P crayon 1.3.4 2017-09-16 [?]
P DBI 1.1.0 2019-12-15 [?]
P dbplyr 1.4.3 2020-04-19 [?]
P digest 0.6.25 2020-02-23 [?]
doParallel 1.0.15 2019-08-02 [1]
P dplyr * 0.8.5 2020-03-07 [?]
P drake 7.12.0 2020-03-25 [?]
P ellipsis 0.3.0 2019-09-20 [?]
P evaluate 0.14 2019-05-28 [?]
P fansi 0.4.1 2020-01-08 [?]
P farver 2.0.3 2020-01-16 [?]
P filelock 1.0.2 2018-10-05 [?]
FNN 1.1.3 2019-02-15 [1]
P forcats * 0.5.0 2020-03-01 [?]
foreach 1.5.0 2020-03-30 [1]
P fs * 1.4.1 2020-04-04 [?]
P future 1.17.0 2020-04-18 [?]
P future.apply 1.5.0 2020-04-17 [?]
gdtools 0.2.2 2020-04-03 [1]
P generics 0.0.2 2018-11-29 [?]
P GetoptLong 0.1.8 2020-01-08 [?]
ggalluvial * 0.12.0 2020-07-14 [1]
P ggplot2 * 3.3.0 2020-03-05 [?]
ggrepel 0.8.2 2020-03-08 [1]
P git2r 0.27.1 2020-05-03 [?]
P GlobalOptions 0.1.1 2019-09-30 [?]
globals 0.12.5 2019-12-07 [1]
P glue * 1.4.0 2020-04-03 [?]
gridBase 0.4-7 2014-02-24 [1]
P gtable 0.3.0 2019-03-25 [?]
P haven 2.2.0 2019-11-08 [?]
P here * 0.1 2017-05-28 [?]
P highr 0.8 2019-03-20 [?]
P hms 0.5.3 2020-01-08 [?]
P htmltools 0.5.0 2020-06-16 [?]
P httpuv 1.5.2 2019-09-11 [?]
P httr 1.4.1 2019-08-05 [?]
P igraph 1.2.5 2020-03-19 [?]
IRanges 2.22.2 2020-05-21 [1]
irlba 2.3.3 2019-02-05 [1]
iterators 1.0.12 2019-07-26 [1]
P jsonlite * 1.6.1 2020-02-02 [?]
P knitr * 1.28 2020-02-06 [?]
P labeling 0.3 2014-08-23 [?]
P later 1.0.0 2019-10-04 [?]
P lattice 0.20-41 2020-04-02 [?]
P lifecycle 0.2.0 2020-03-06 [?]
listenv 0.8.0 2019-12-05 [1]
P lubridate 1.7.8 2020-04-06 [?]
P magrittr 1.5 2014-11-22 [?]
P Matrix 1.2-18 2019-11-27 [?]
P memoise 1.1.0 2017-04-21 [?]
P modelr 0.1.7 2020-04-30 [?]
P munsell 0.5.0 2018-06-12 [?]
network 1.16.0 2019-12-01 [1]
P nlme 3.1-147 2020-04-13 [?]
NMF 0.22.0 2020-02-12 [1]
P pander * 0.6.3 2018-11-06 [?]
pbapply 1.4-2 2019-08-31 [1]
P pillar 1.4.4 2020-05-05 [?]
P pkgconfig 2.0.3 2019-09-22 [?]
pkgmaker 0.31.1 2020-03-19 [1]
P plyr 1.8.6 2020-03-03 [?]
png 0.1-7 2013-12-03 [1]
P prettyunits 1.1.1 2020-01-24 [?]
P progress 1.2.2 2019-05-16 [?]
P promises 1.1.0 2019-10-04 [?]
P purrr * 0.3.4 2020-04-17 [?]
P R6 2.4.1 2019-11-12 [?]
P RColorBrewer 1.1-2 2014-12-07 [?]
P Rcpp 1.0.4.6 2020-04-09 [?]
P readr * 1.3.1 2018-12-21 [?]
P readxl 1.3.1 2019-03-13 [?]
registry 0.5-1 2019-03-05 [1]
P renv 0.12.0 2020-08-28 [?]
P repr 1.1.0 2020-01-28 [?]
P reprex 0.3.0 2019-05-16 [?]
P reshape2 1.4.4 2020-04-09 [?]
P reticulate 1.16 2020-05-27 [?]
rjson 0.2.20 2018-06-08 [1]
P rlang 0.4.6 2020-05-02 [?]
P rmarkdown 2.1 2020-01-20 [?]
rngtools 1.5 2020-01-23 [1]
P rprojroot 1.3-2 2018-01-03 [?]
RSpectra 0.16-0 2019-12-01 [1]
P rstudioapi 0.11 2020-02-07 [?]
P rvest 0.3.5 2019-11-08 [?]
S4Vectors 0.26.1 2020-05-16 [1]
P scales 1.1.0 2019-11-18 [?]
P sessioninfo 1.1.1 2018-11-05 [?]
P shape 1.4.4 2018-02-07 [?]
P skimr * 2.1.1 2020-04-16 [?]
sna 2.5 2019-12-10 [1]
statnet.common 4.3.0 2019-06-02 [1]
P storr 1.2.1 2018-10-18 [?]
P stringi 1.4.6 2020-02-17 [?]
P stringr * 1.4.0 2019-02-10 [?]
svglite 1.2.3.2 2020-07-07 [1]
systemfonts 0.2.3 2020-06-09 [1]
P tibble * 3.0.1 2020-04-20 [?]
P tidyr * 1.0.3 2020-05-07 [?]
P tidyselect 1.0.0 2020-01-27 [?]
P tidyverse * 1.3.0 2019-11-21 [?]
P txtq 0.2.0 2019-10-15 [?]
P vctrs 0.2.4 2020-03-10 [?]
P whisker 0.4 2019-08-28 [?]
P withr 2.2.0 2020-04-20 [?]
P workflowr 1.6.2 2020-04-30 [?]
P xfun 0.13 2020-04-13 [?]
P xml2 1.3.2 2020-04-23 [?]
xtable 1.8-4 2019-04-21 [1]
P yaml 2.2.1 2020-02-01 [?]
source
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
standard (@1.4)
CRAN (R 4.0.2)
Bioconductor
Bioconductor
CRAN (R 4.0.0)
Github (sqjin/CellChat@1df06c1)
standard (@1.1.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.2)
CRAN (R 4.0.2)
standard (@1.4-1)
Github (jokergoo/ComplexHeatmap@d38254f)
standard (@1.0.4)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.2)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
standard (@0.14)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.1)
CRAN (R 4.0.0)
CRAN (R 4.0.2)
standard (@0.0.2)
CRAN (R 4.0.0)
CRAN (R 4.0.2)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.2)
standard (@0.3.0)
standard (@2.2.0)
standard (@0.1)
standard (@0.8)
CRAN (R 4.0.0)
CRAN (R 4.0.2)
standard (@1.5.2)
standard (@1.4.1)
CRAN (R 4.0.0)
Bioconductor
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
standard (@0.3)
standard (@1.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
standard (@1.2-18)
standard (@1.1.0)
CRAN (R 4.0.0)
standard (@0.5.0)
CRAN (R 4.0.2)
CRAN (R 4.0.0)
CRAN (R 4.0.2)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.2)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
standard (@1.1.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
standard (@1.1-2)
CRAN (R 4.0.0)
standard (@1.3.1)
standard (@1.3.1)
CRAN (R 4.0.2)
CRAN (R 4.0.2)
CRAN (R 4.0.0)
standard (@0.3.0)
CRAN (R 4.0.0)
CRAN (R 4.0.2)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.2)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
standard (@0.3.5)
Bioconductor
standard (@1.1.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.2)
CRAN (R 4.0.2)
standard (@1.2.1)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.2)
CRAN (R 4.0.2)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
standard (@1.3.0)
standard (@0.2.0)
CRAN (R 4.0.0)
standard (@0.4)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.2)
CRAN (R 4.0.0)
[1] /Users/luke.zappia/Documents/Projects/interaction-tools/renv/library/R-4.0/x86_64-apple-darwin17.0
[2] /private/var/folders/rj/60lhr791617422kqvh0r4vy40000gn/T/RtmpOqMPdA/renv-system-library
[3] /private/var/folders/rj/60lhr791617422kqvh0r4vy40000gn/T/RtmpqYMqtc/renv-system-library
[4] /private/var/folders/rj/60lhr791617422kqvh0r4vy40000gn/T/Rtmpdw6ROp/renv-system-library
P ── Loaded and on-disk path mismatch.Chunk time: 0.31 secs