Method: MNN
This page displays information for the following method:
Name: |
MNN |
Description: |
Mutual nearest neighbours |
GitHub: |
|
DOI: |
|
Labels required?: |
No |
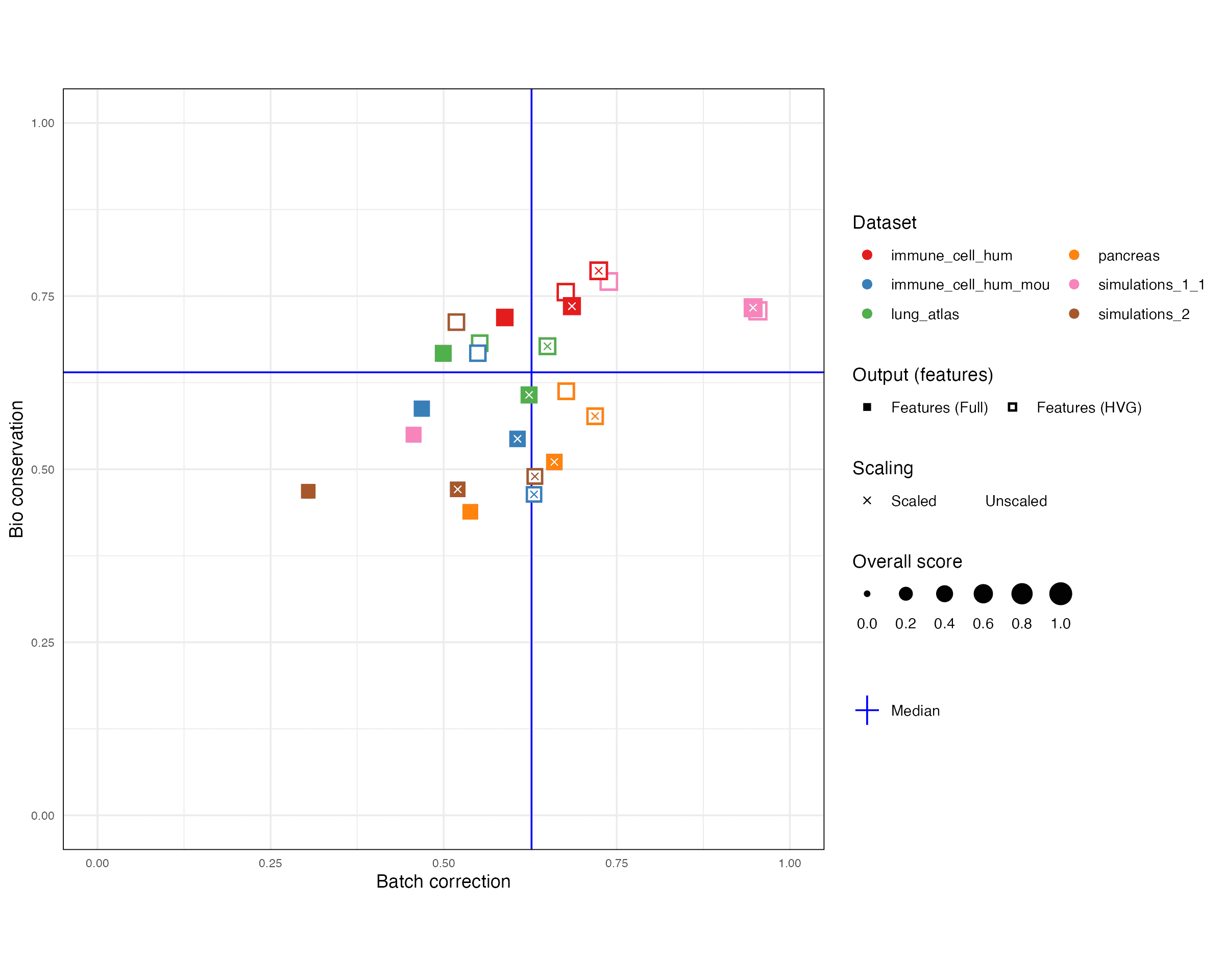
1 Overall
1.1 Plot
1.2 Table
2 Individual metrics
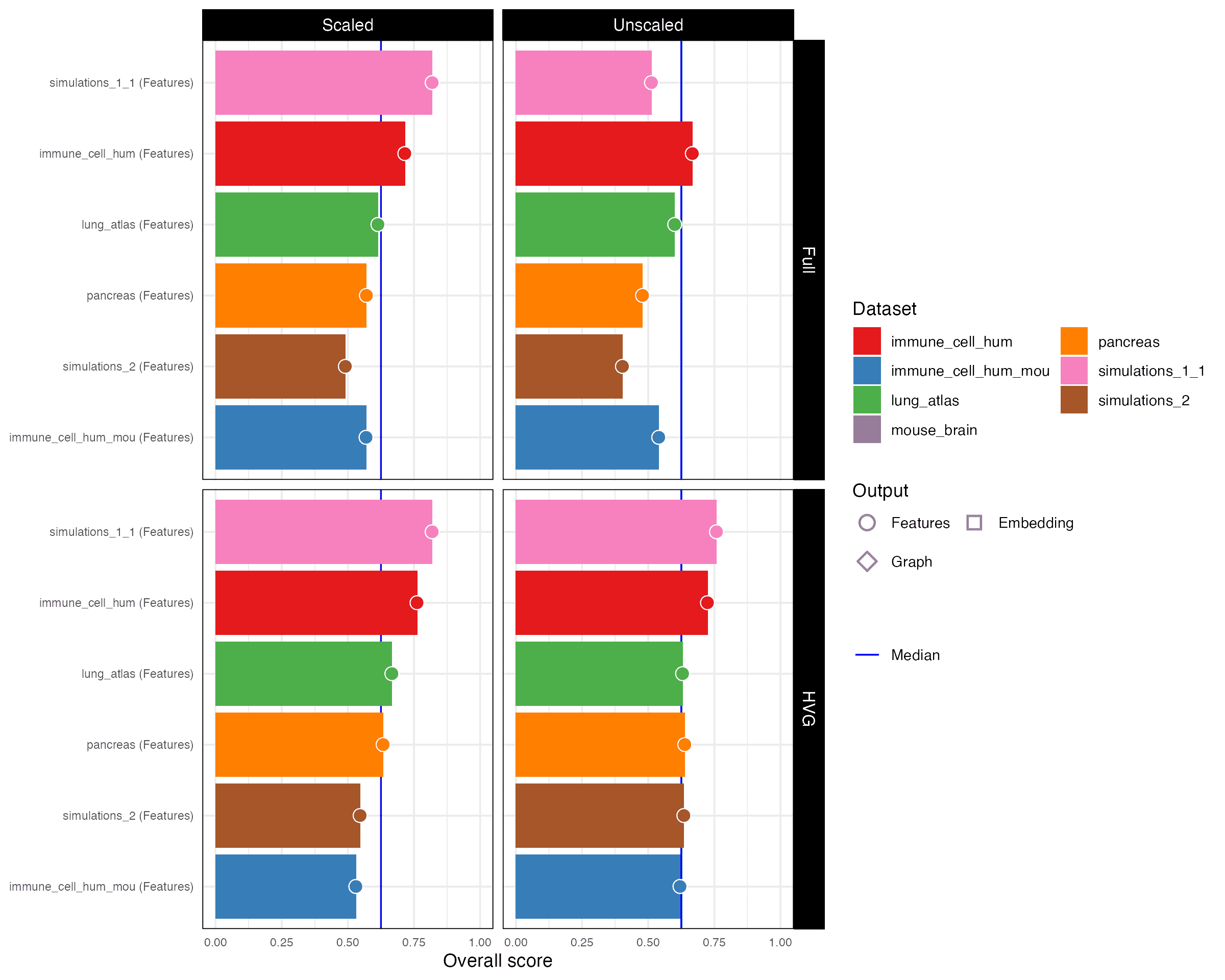
2.1 Overall
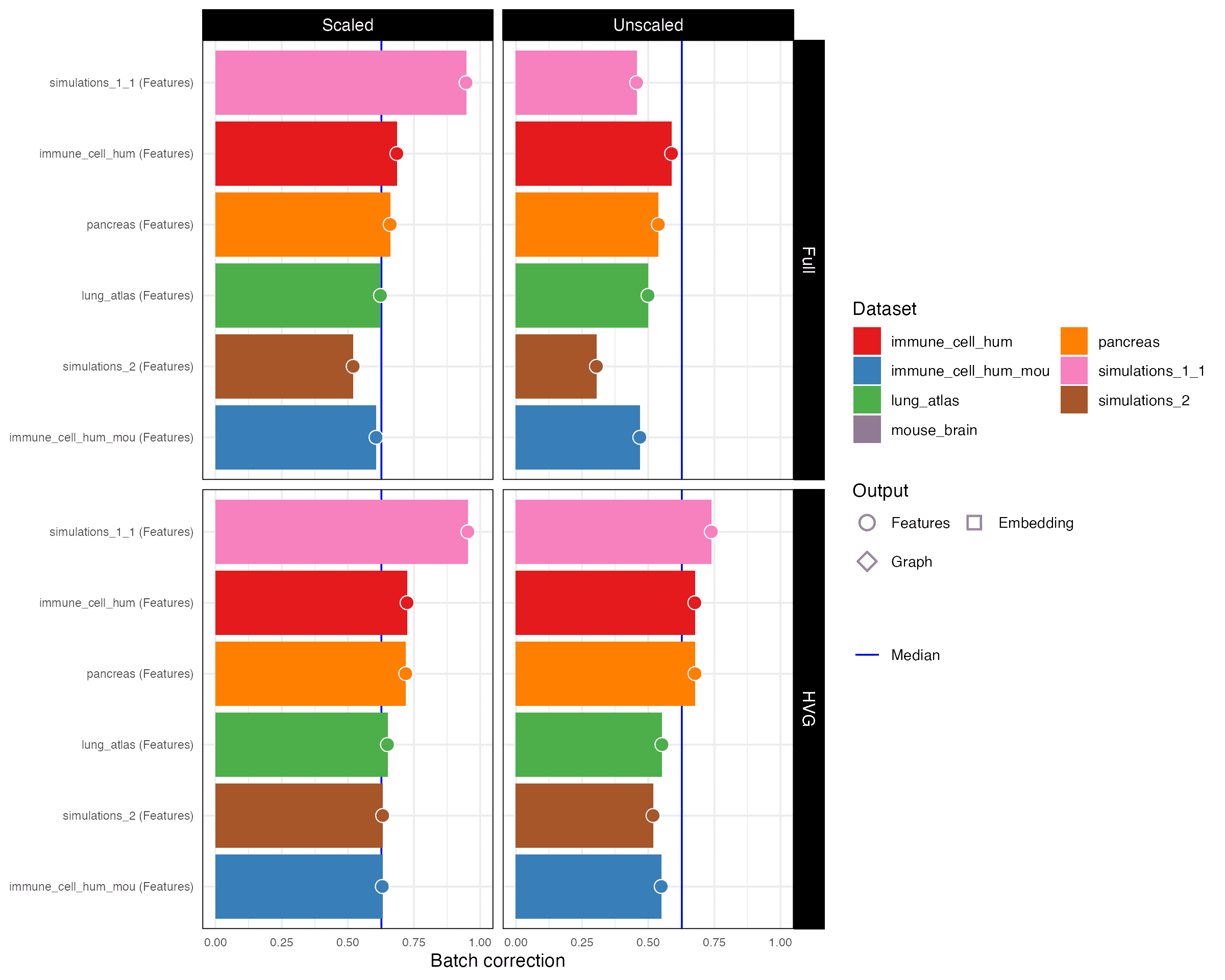
2.2 Batch correction
Batch correction
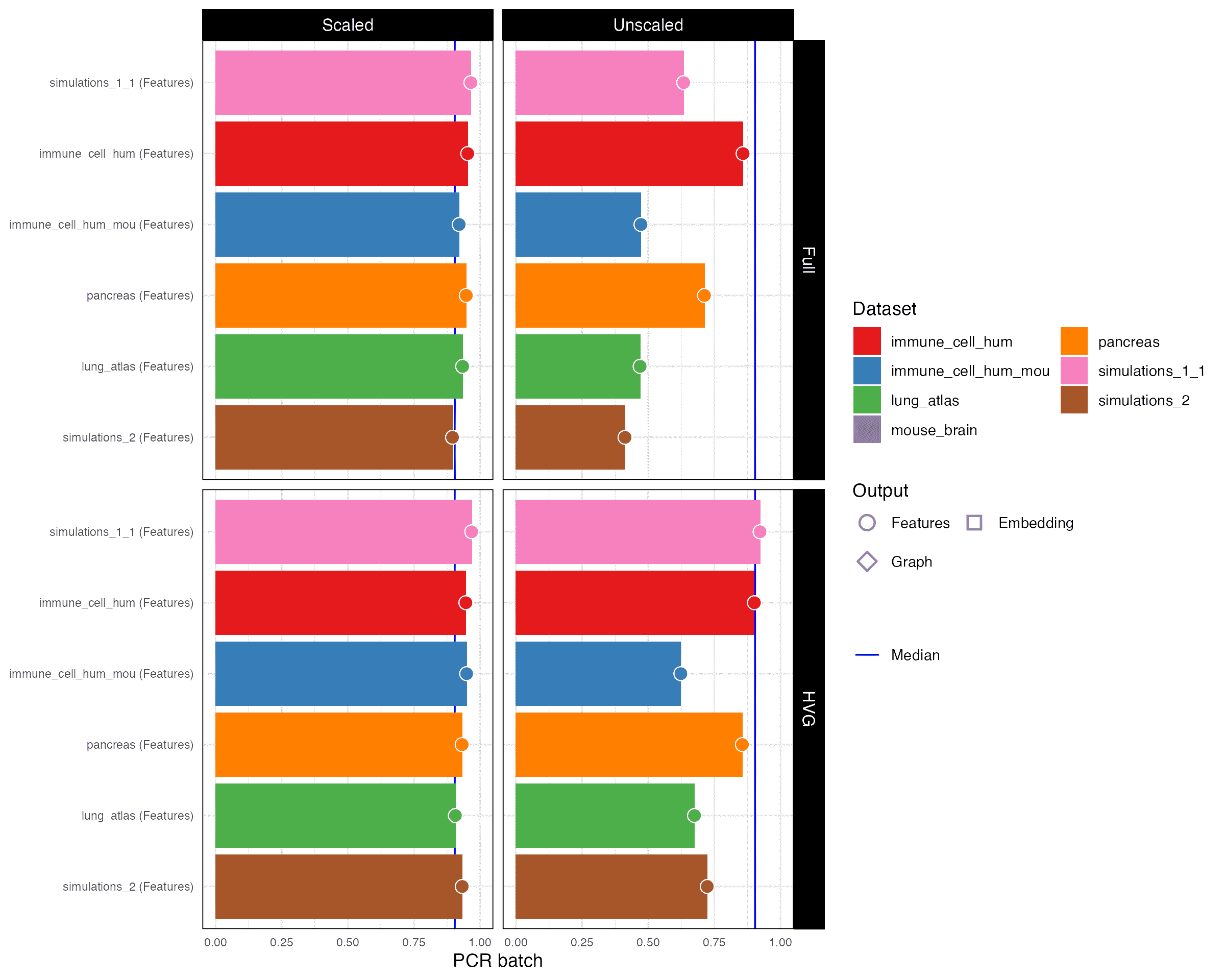
PCR batch
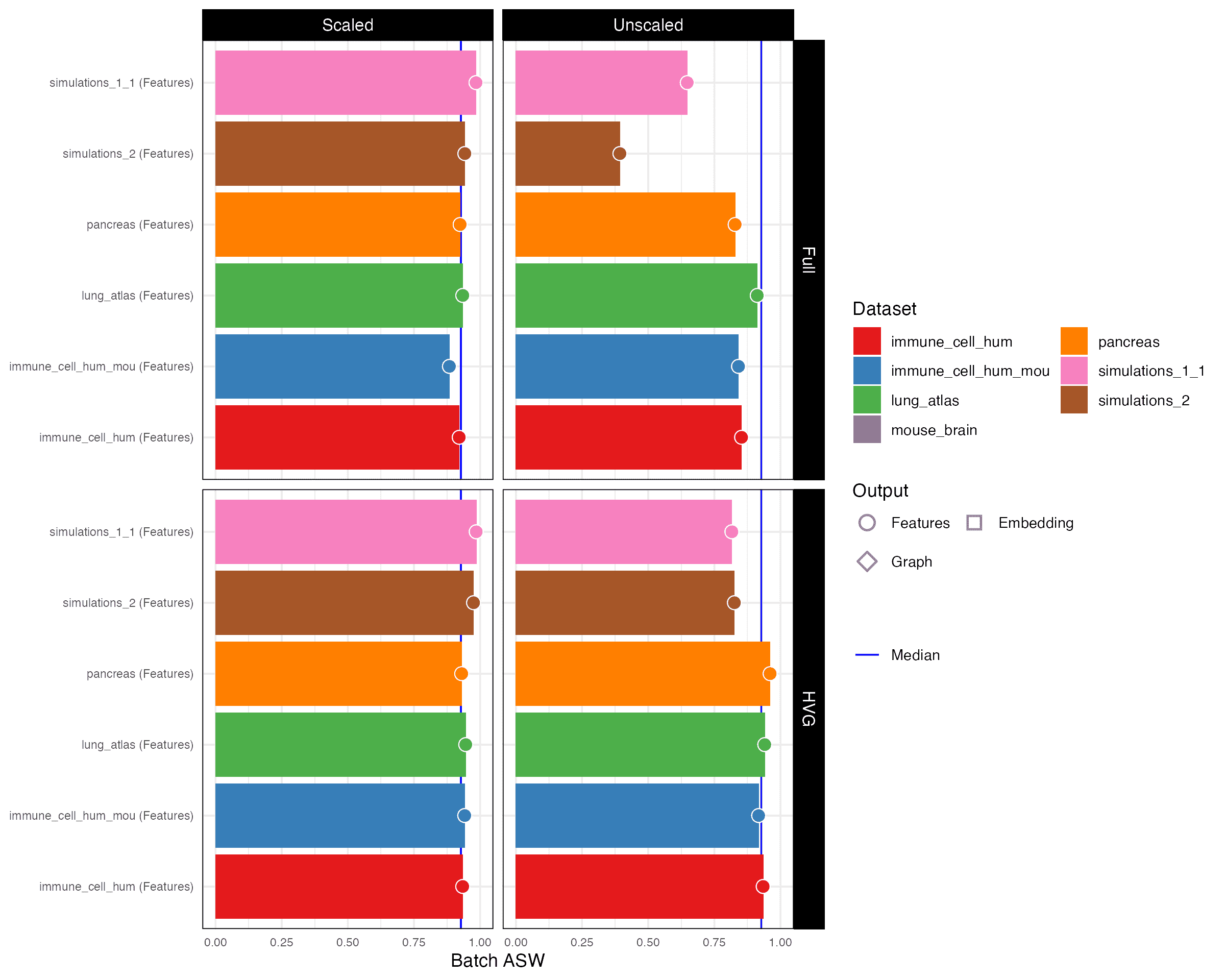
Batch ASW
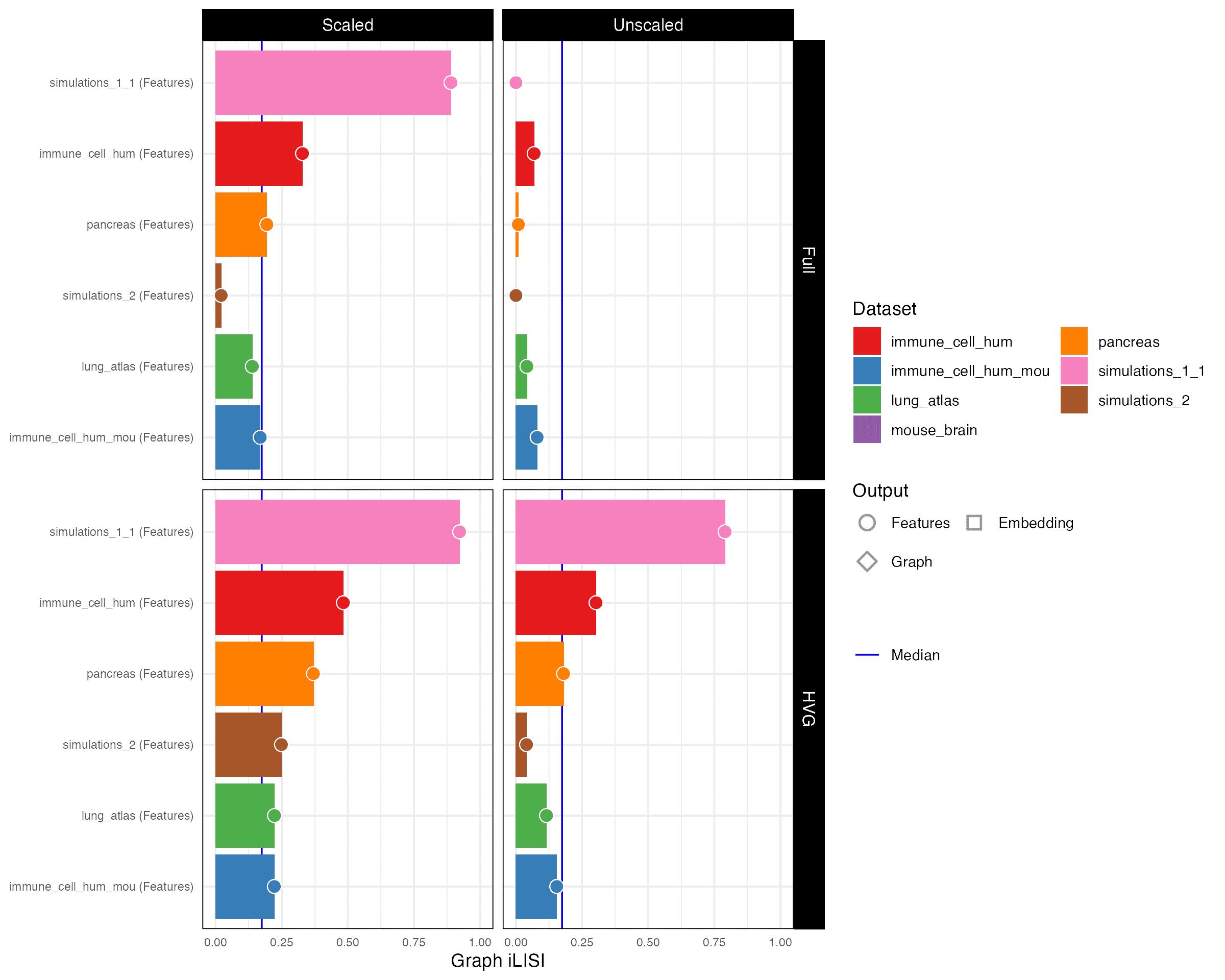
Graph iLISI
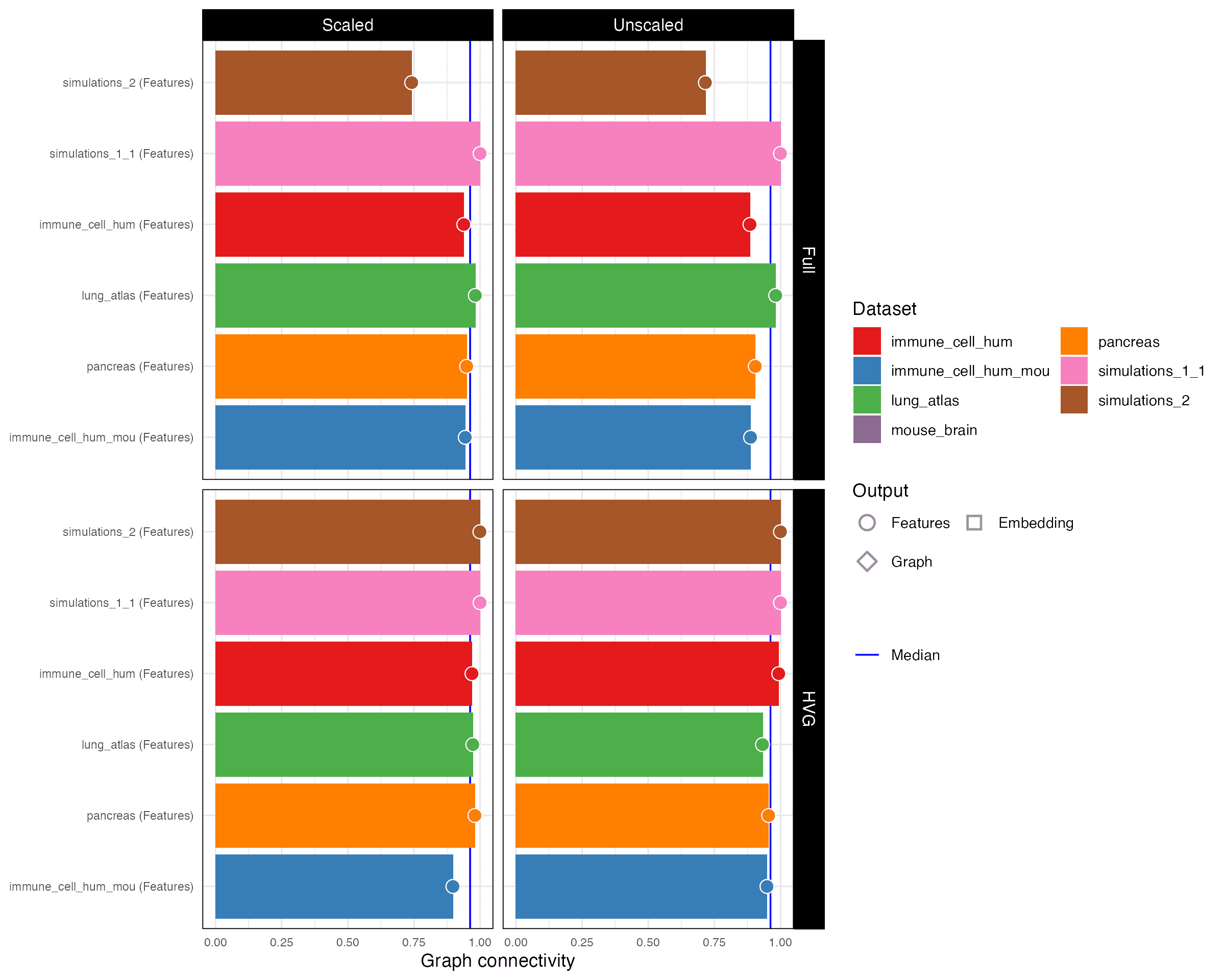
Graph connectivity
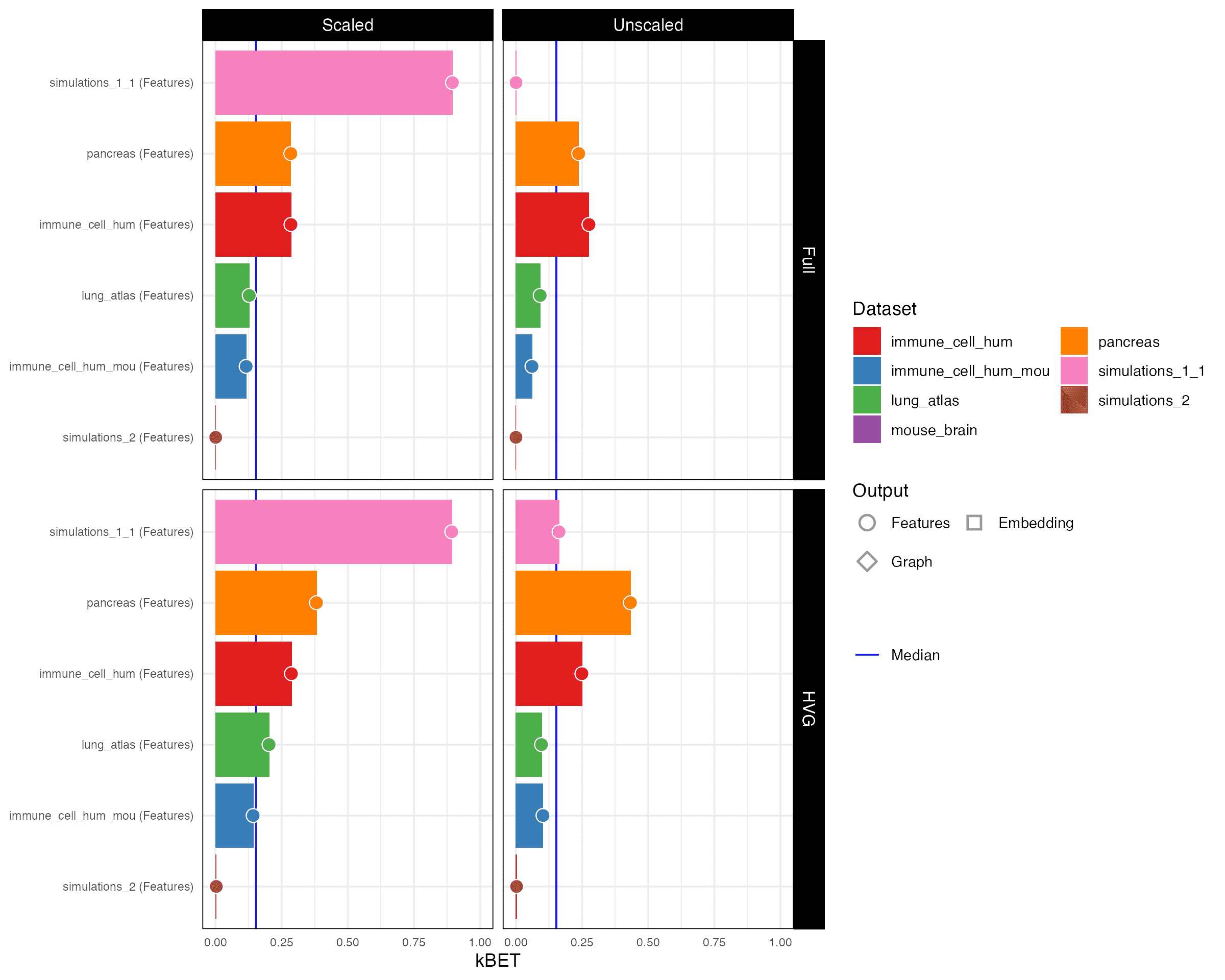
kBET
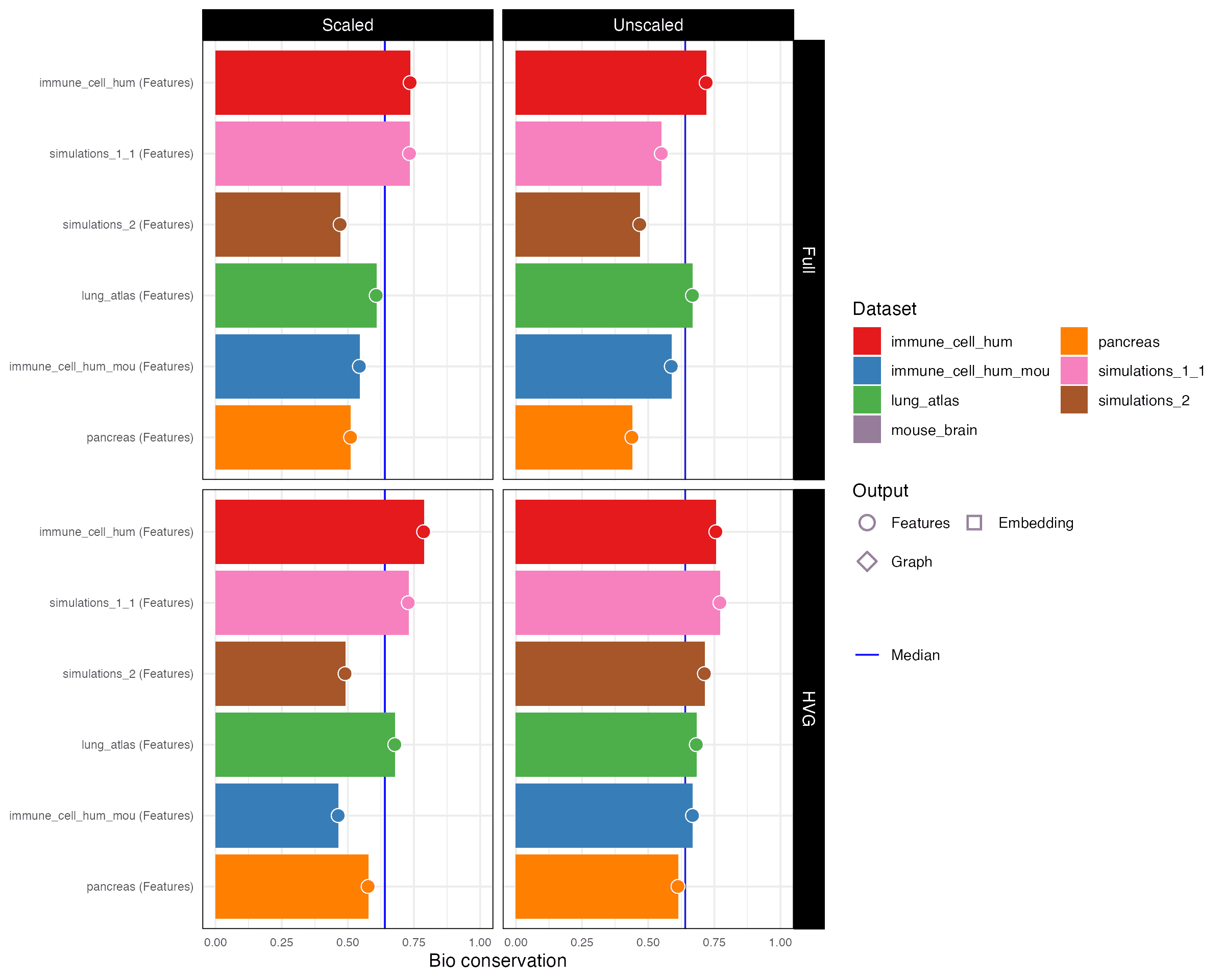
2.3 Bio conservation
Bio conservation
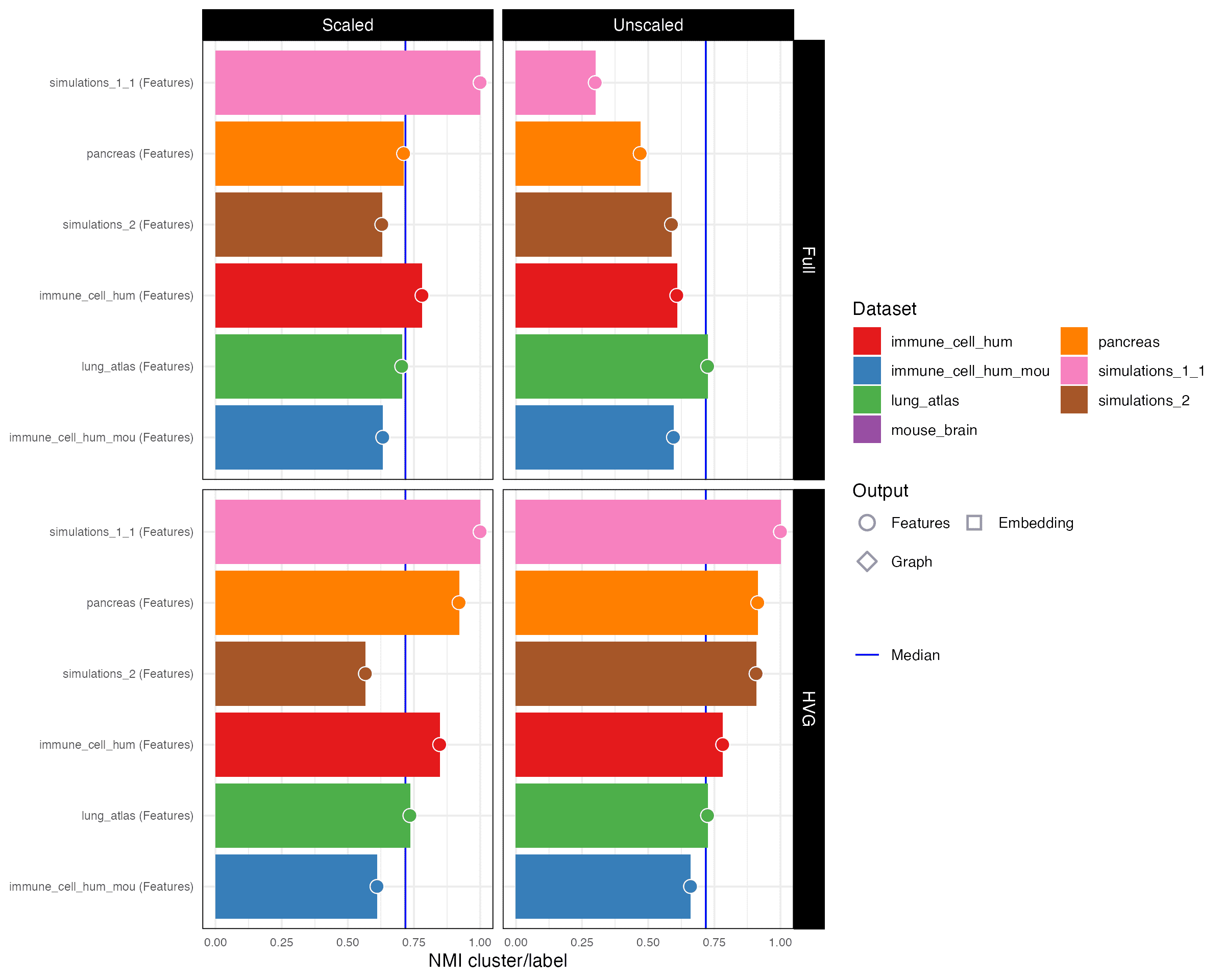
NMI cluster/label
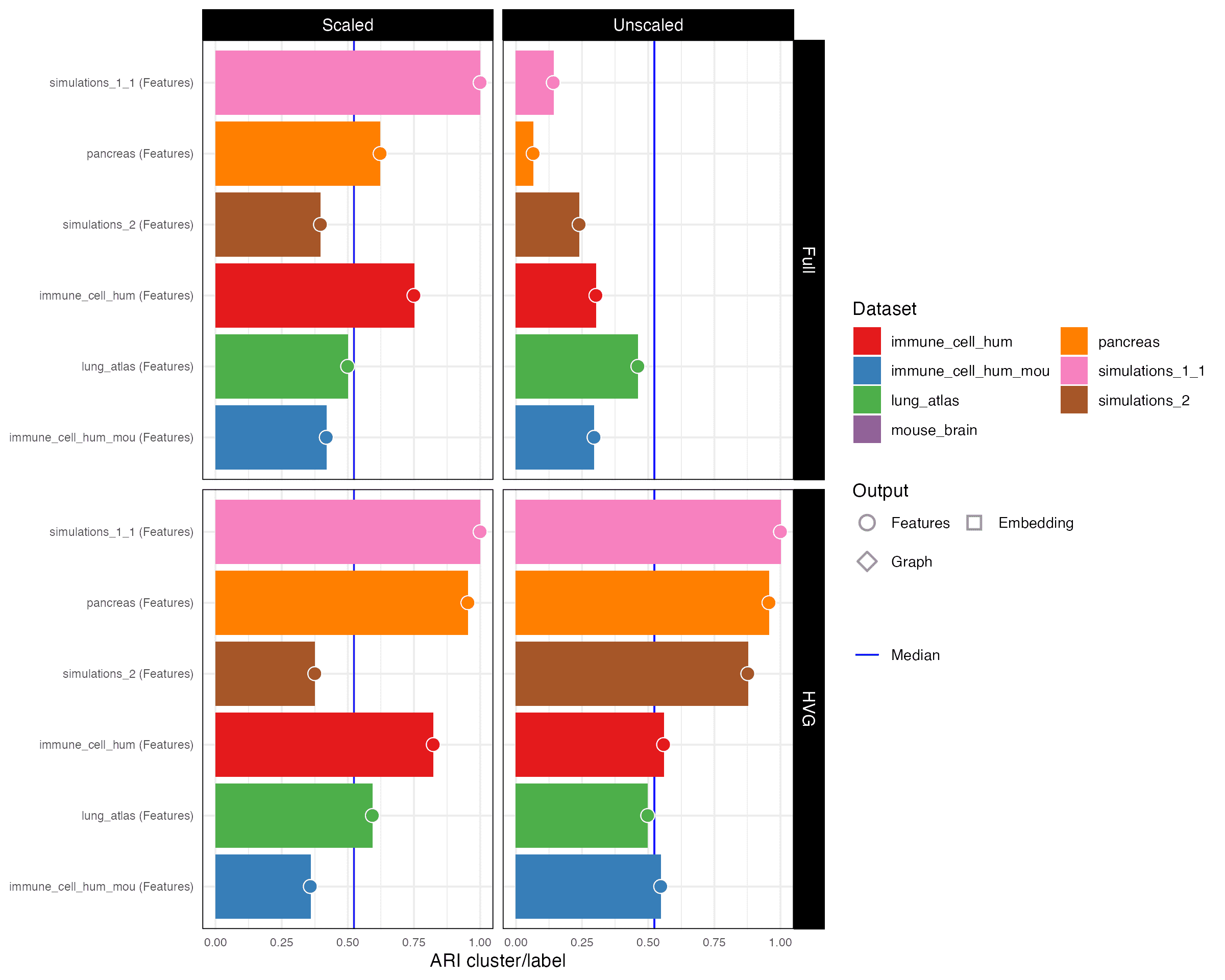
ARI cluster/label
Label ASW
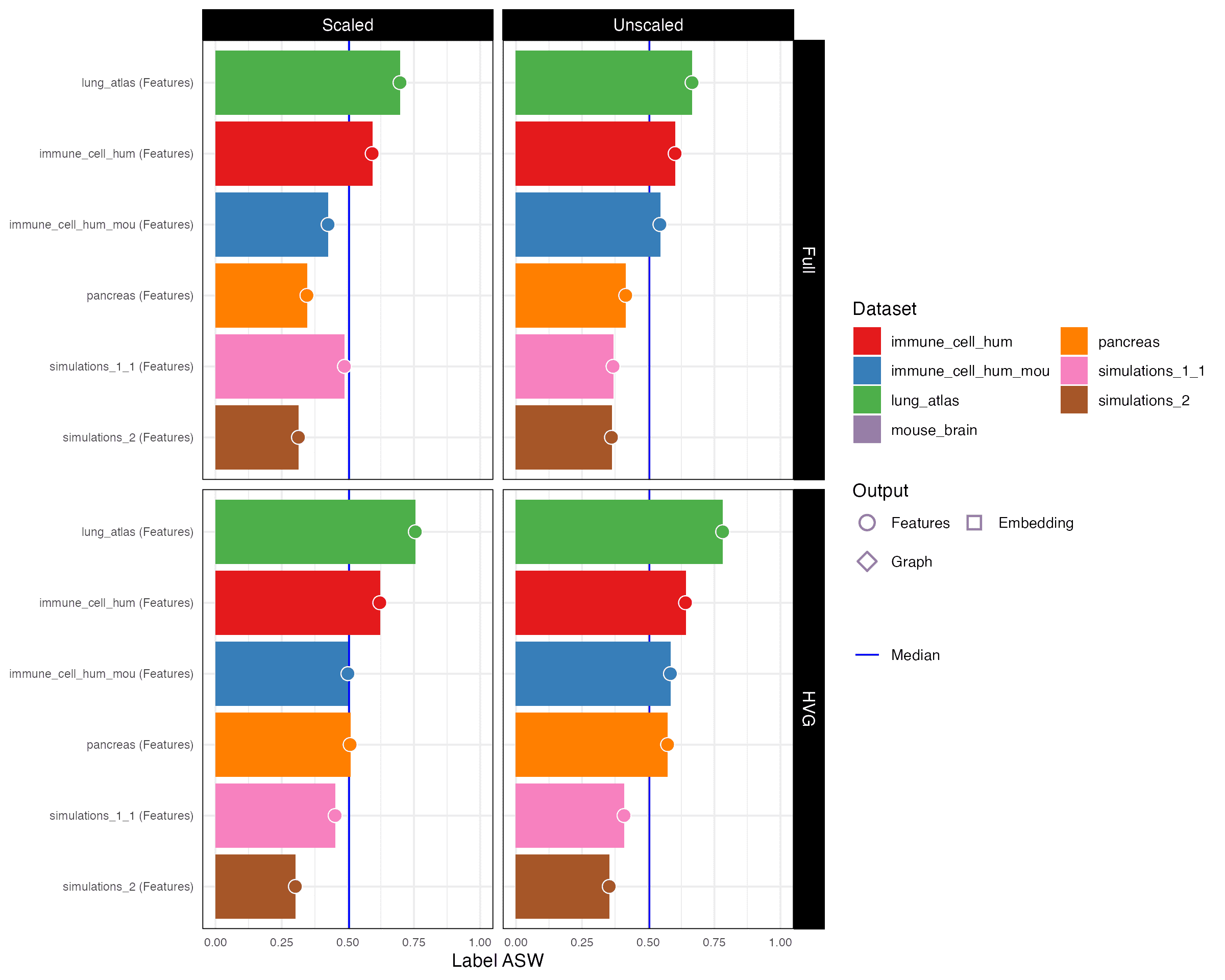
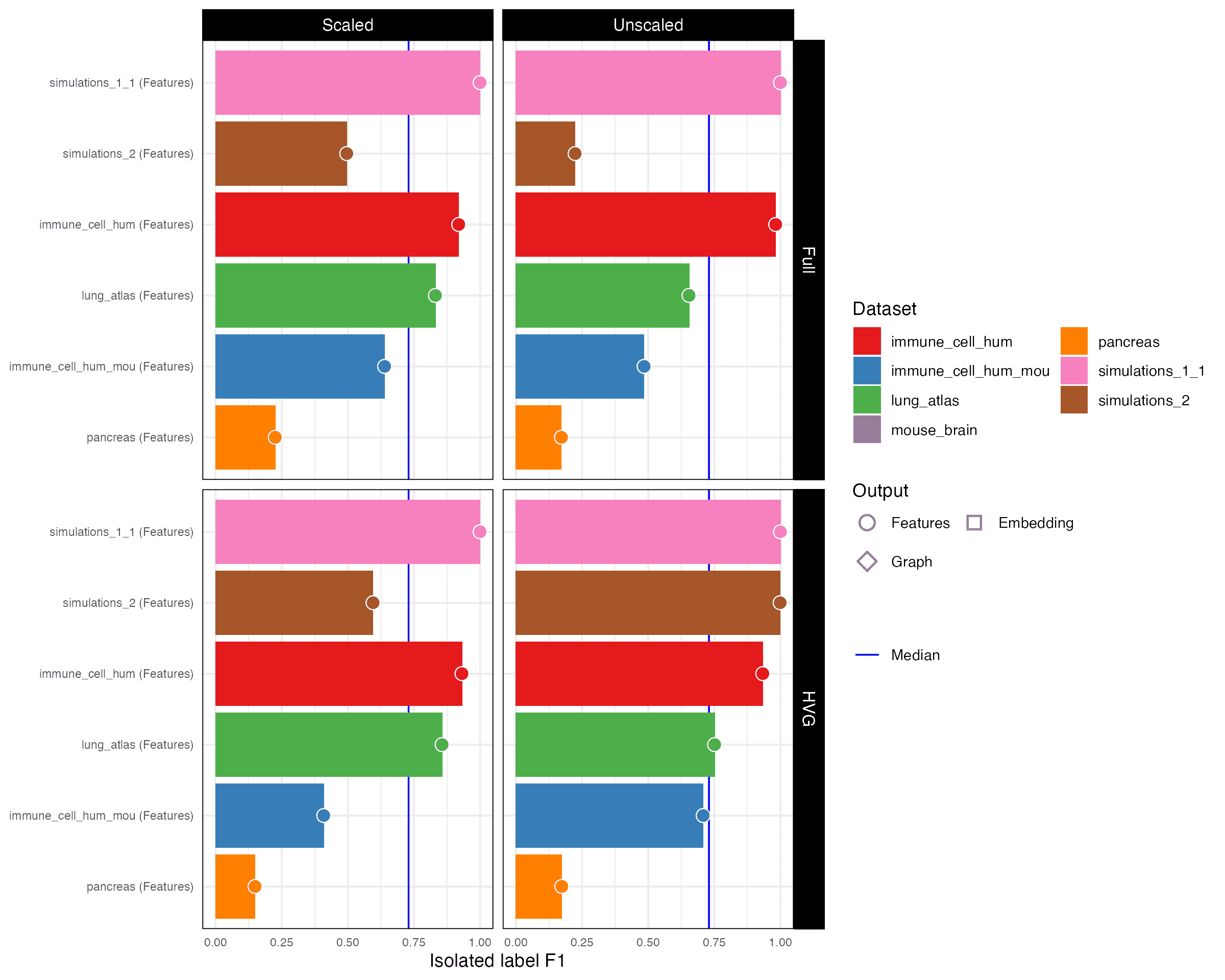
Isolated label F1
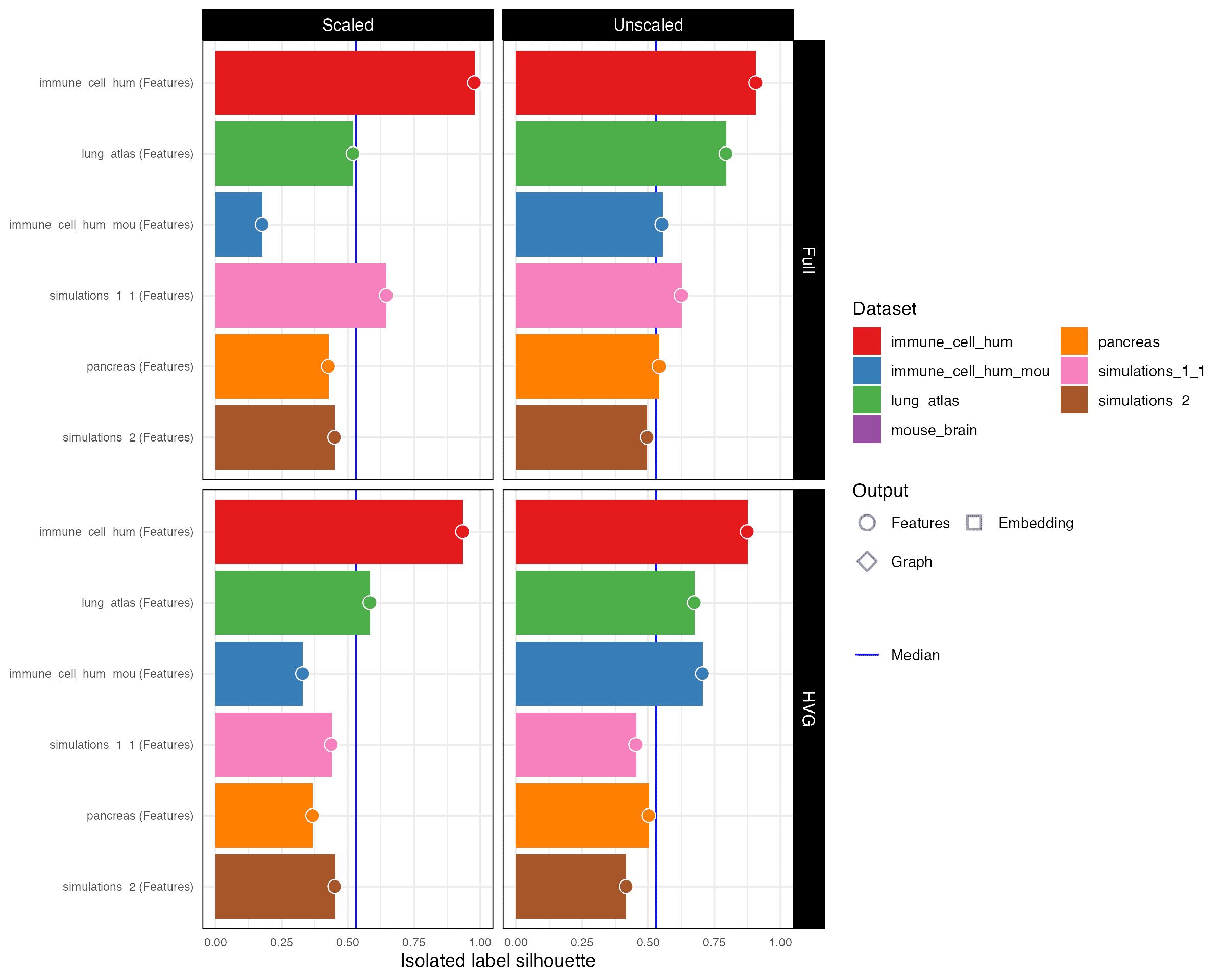
Isolated label silhouette
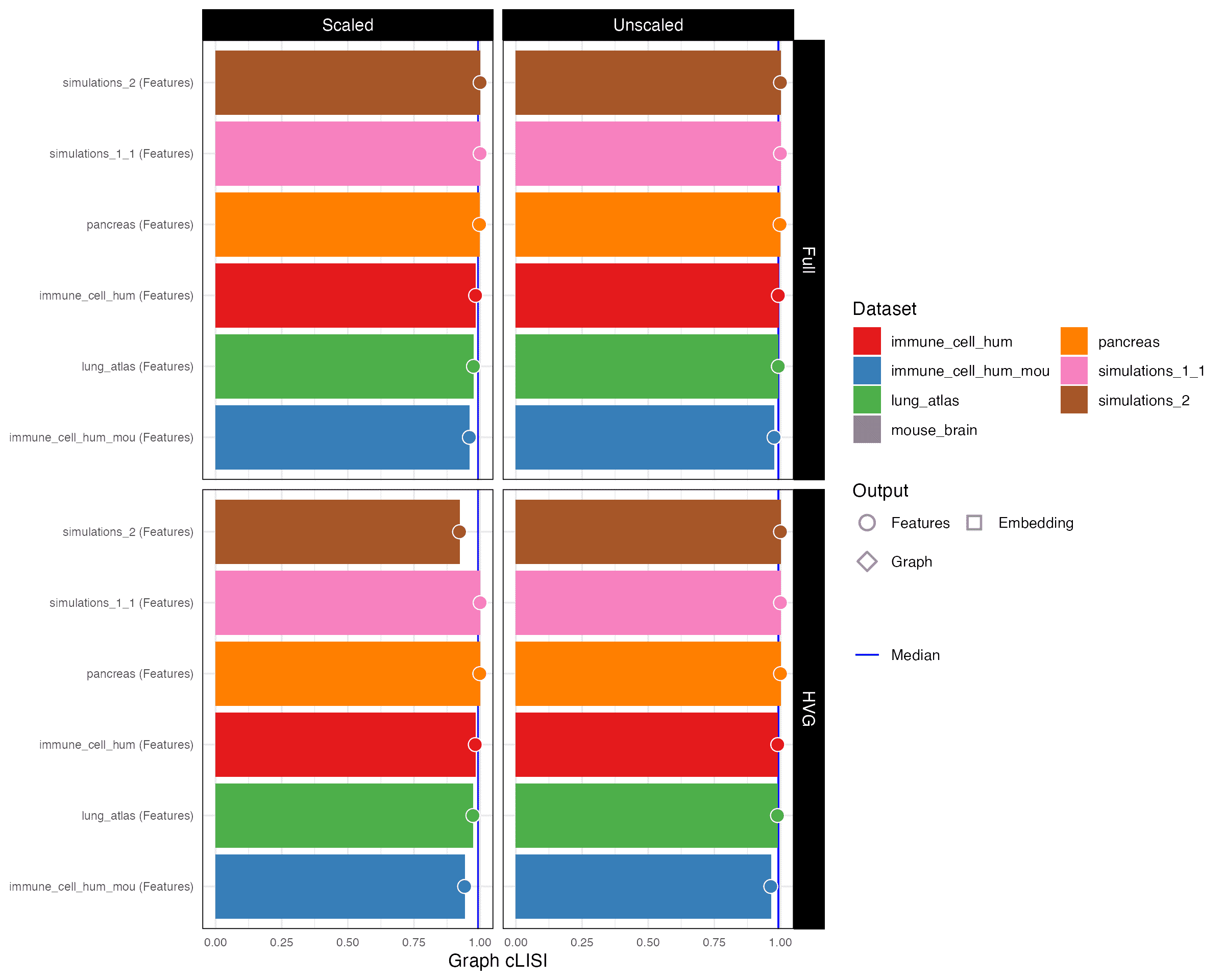
Graph cLISI
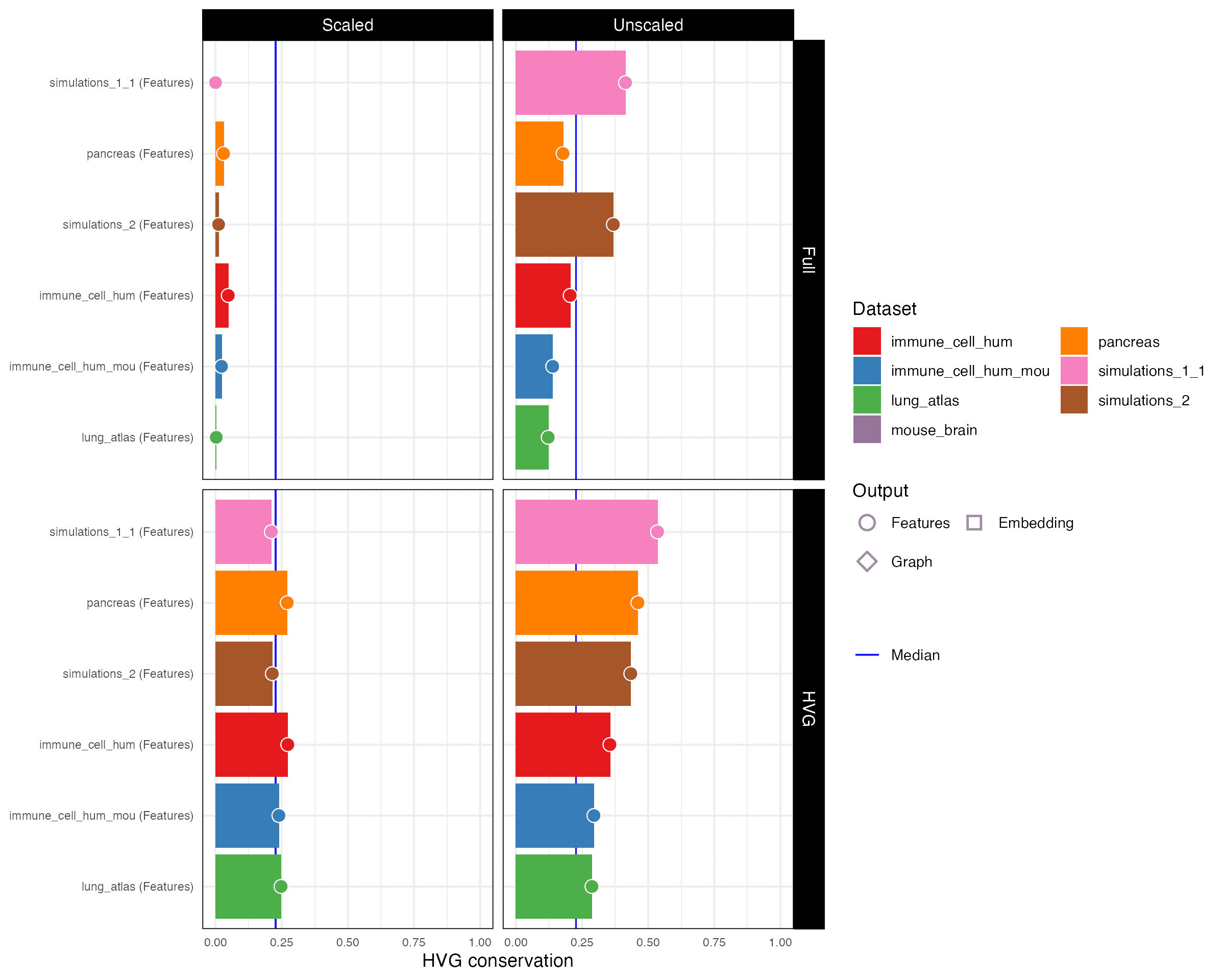
HVG conservation
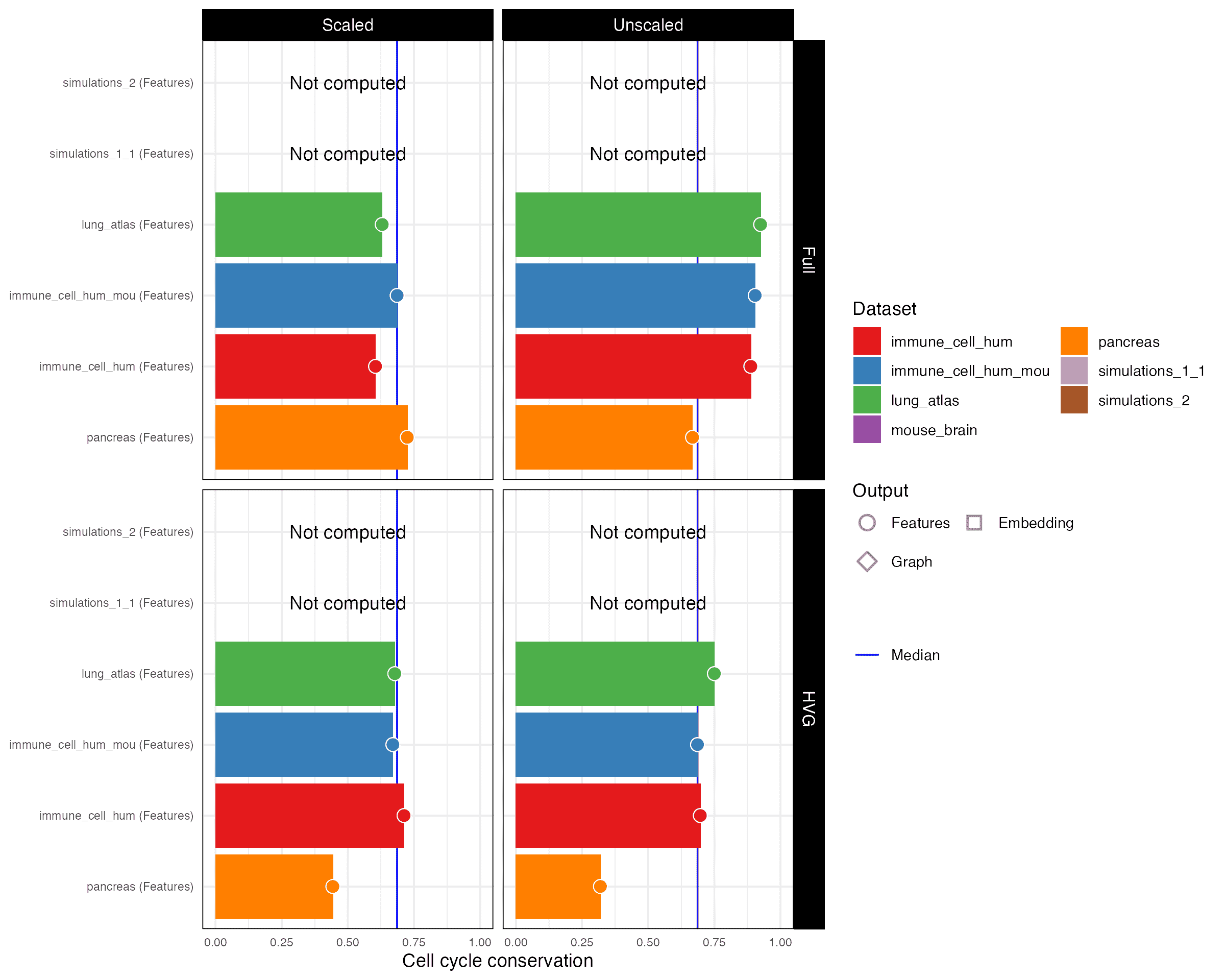
Cell cycle conservation
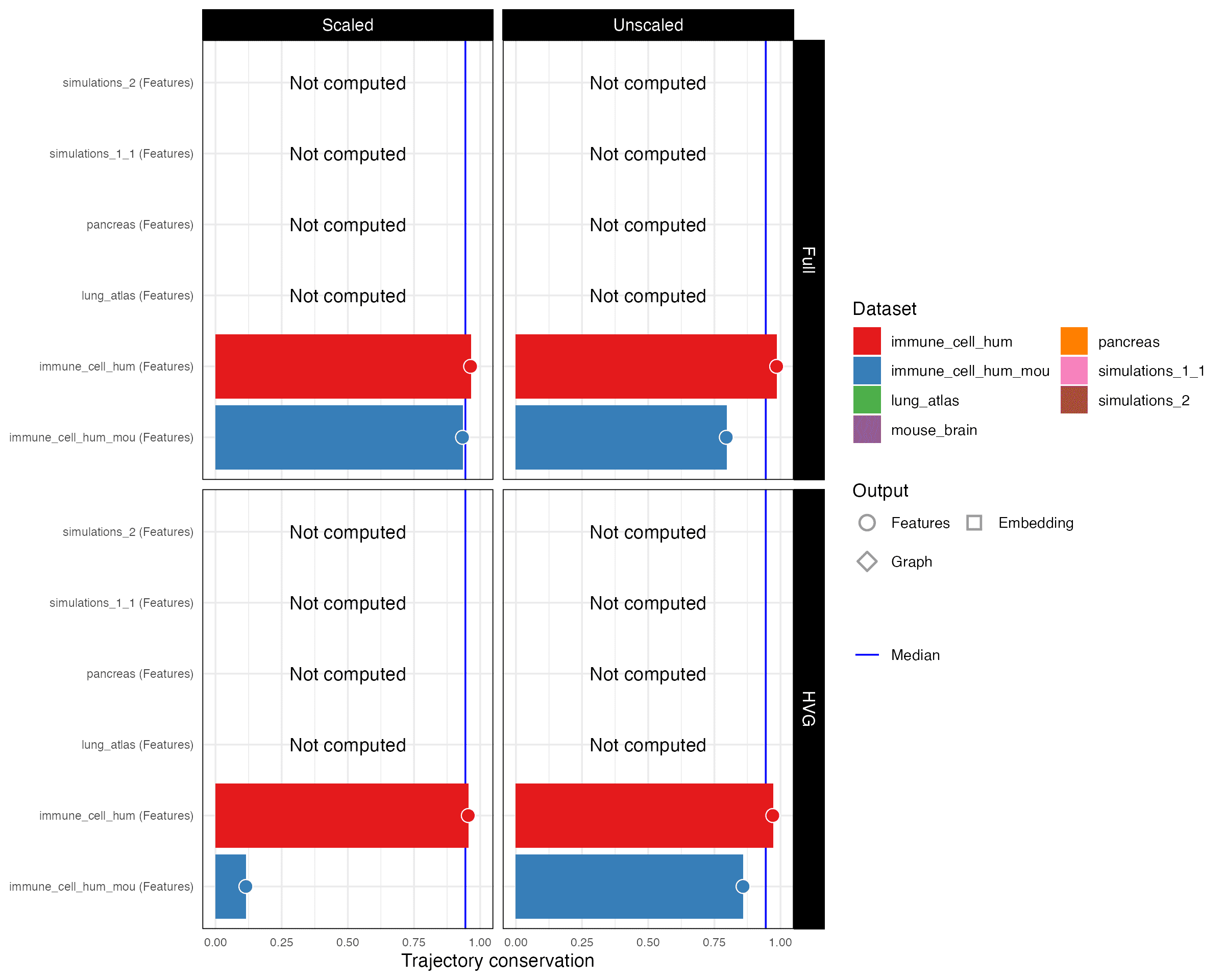
Trajectory conservation
3 Embeddings
Dataset (Output)
simulations_1_1 (Features)
Input features (Scaling)
Full (Scaled)
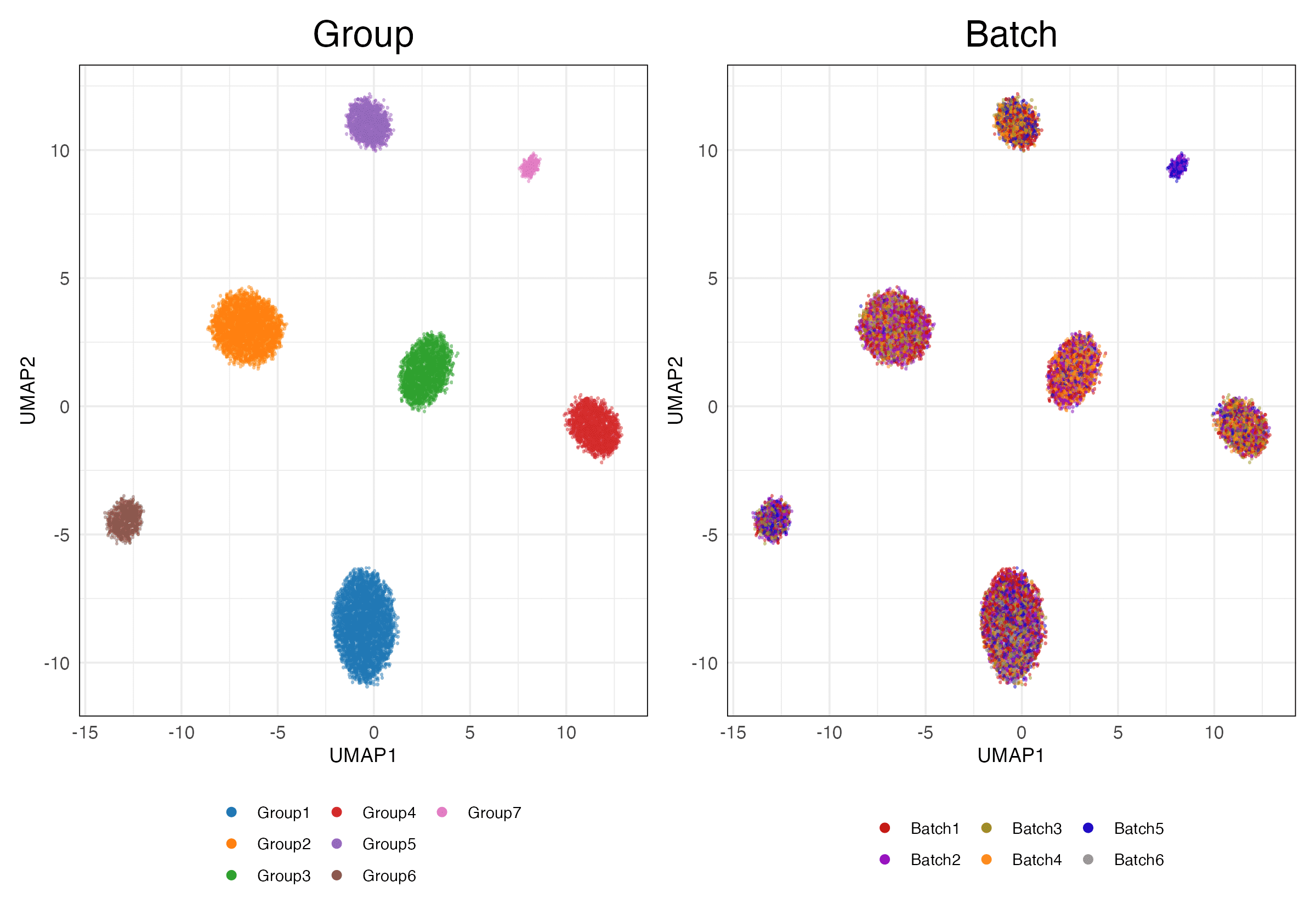
Full (Unscaled)
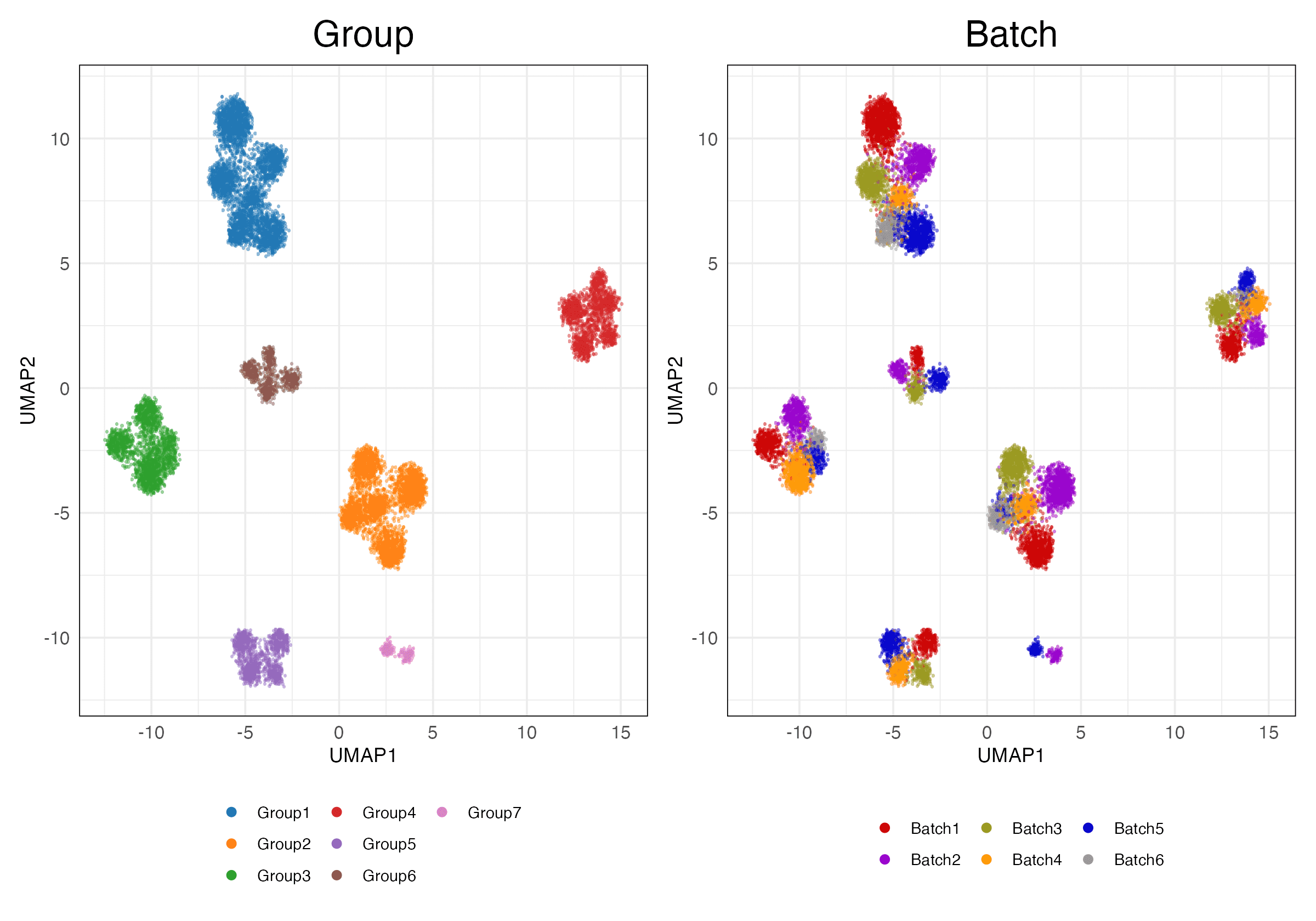
HVG (Scaled)
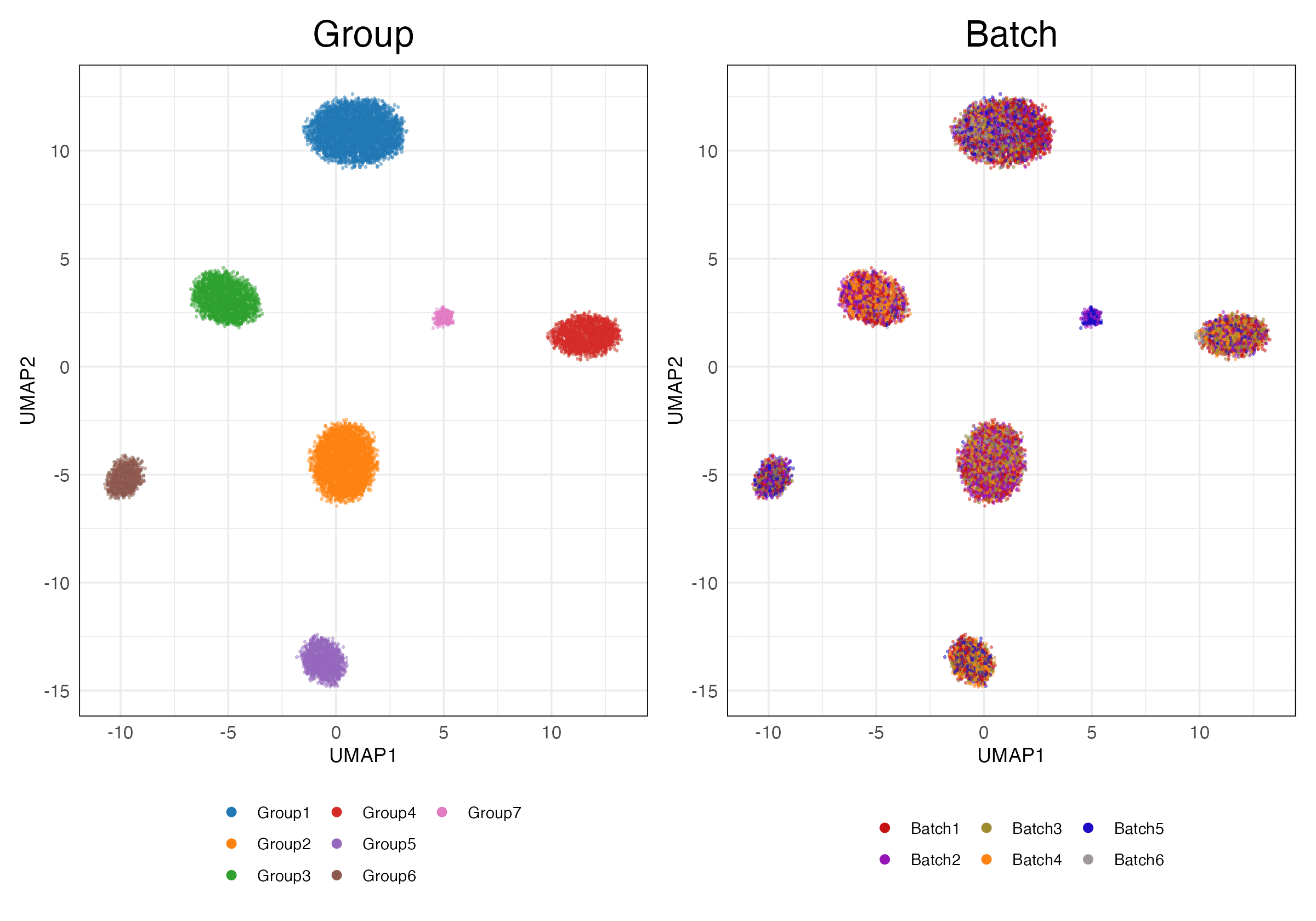
HVG (Unscaled)
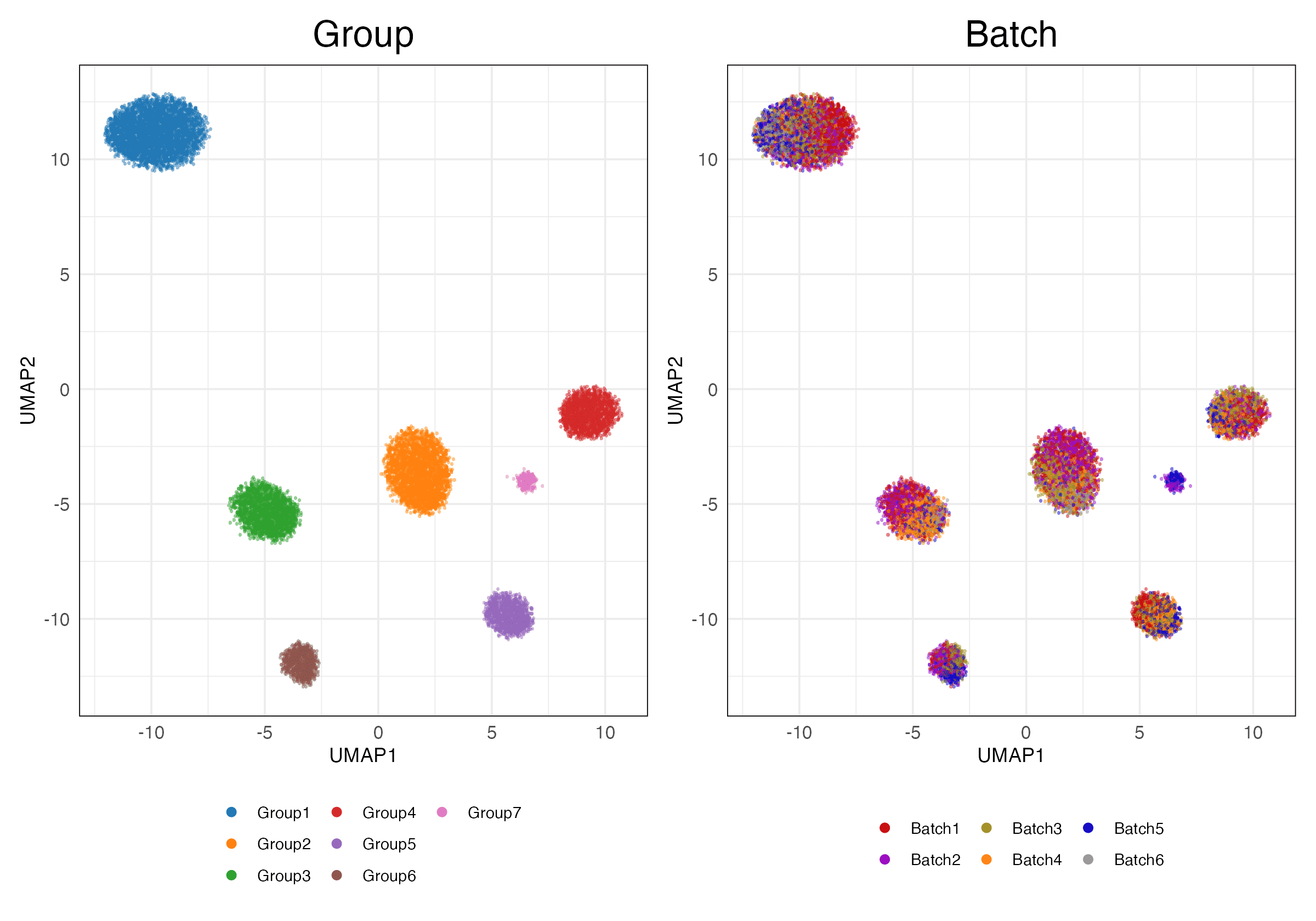
immune_cell_hum (Features)
Input features (Scaling)
Full (Scaled)
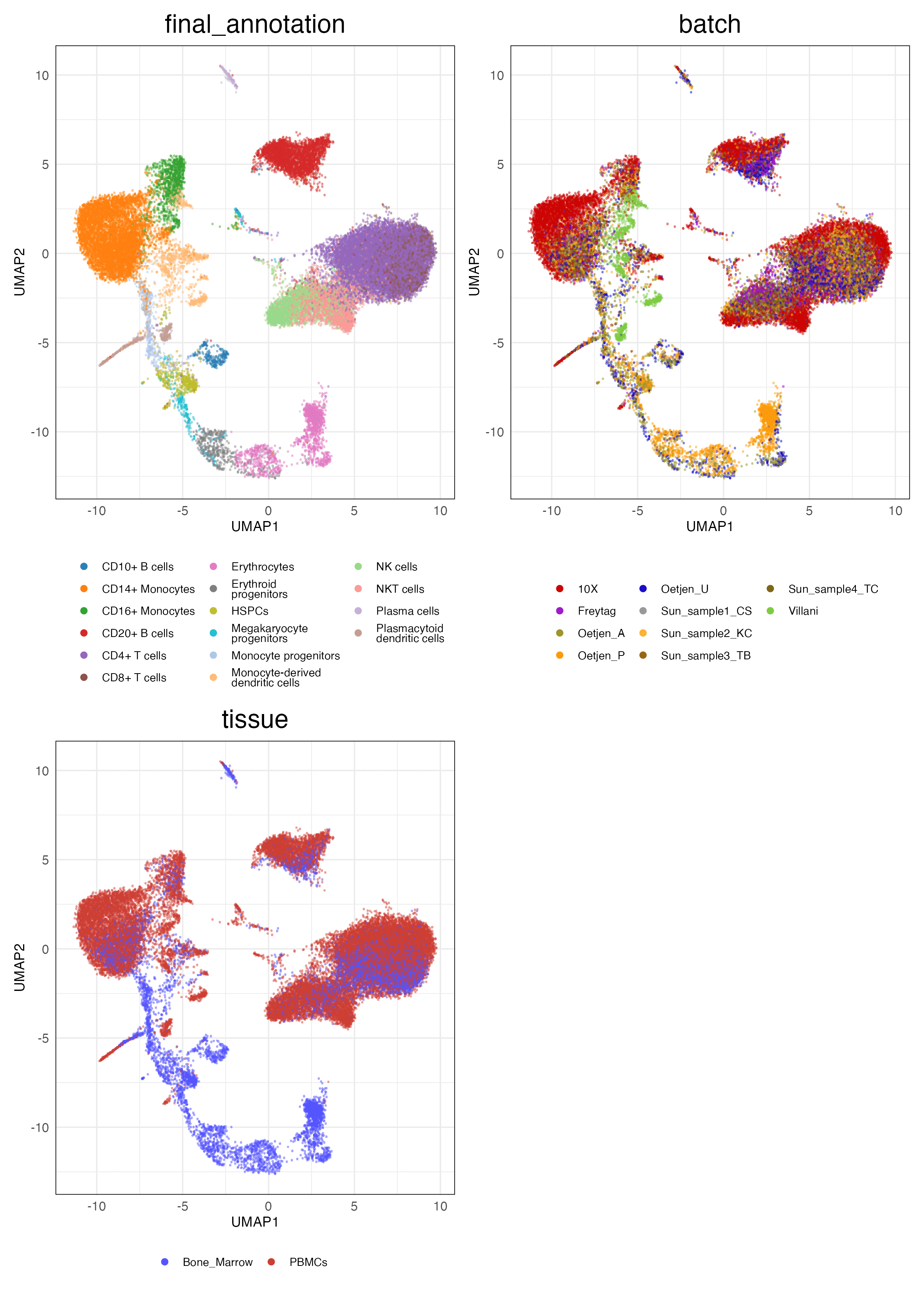
Full (Unscaled)
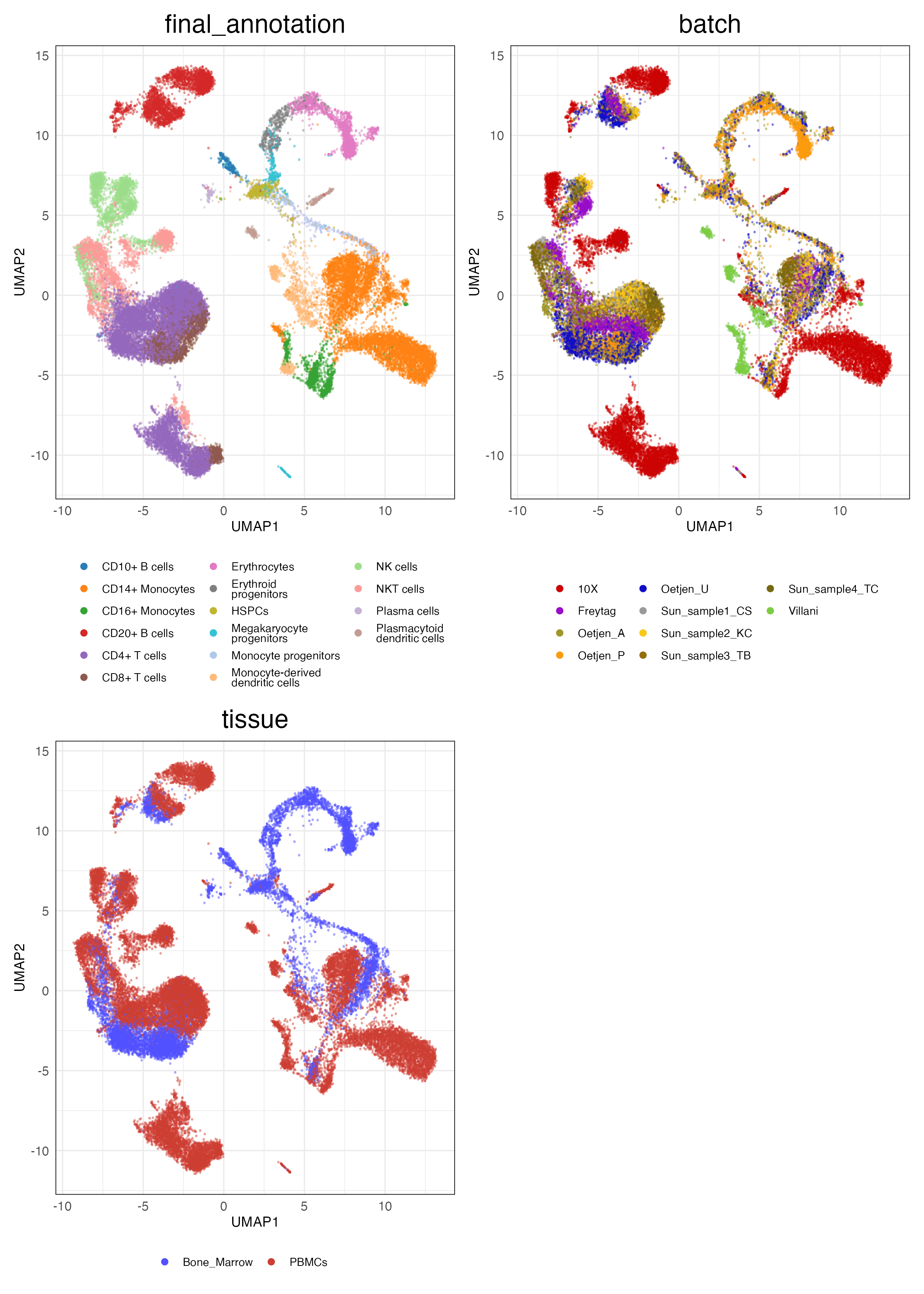
HVG (Scaled)
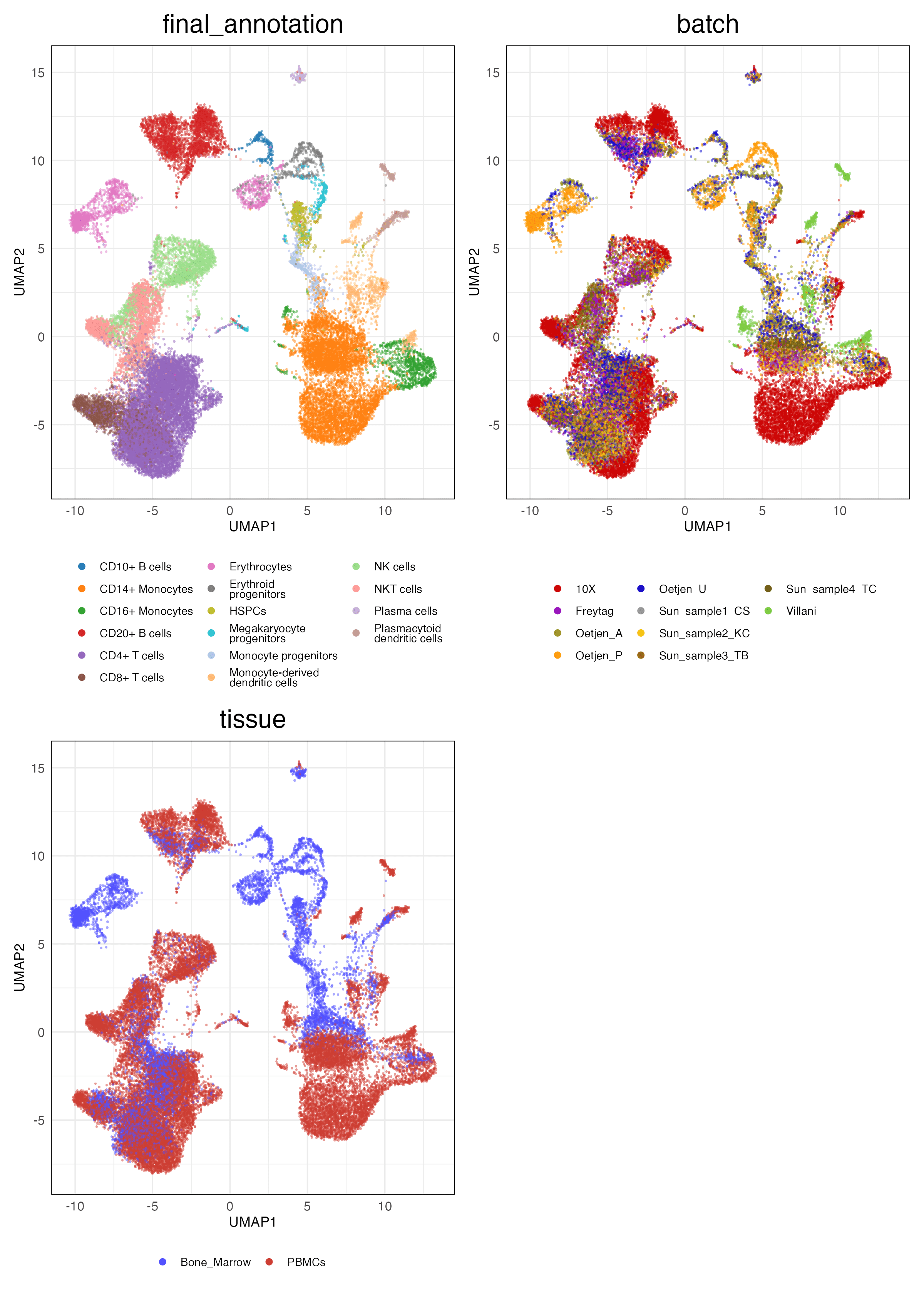
HVG (Unscaled)
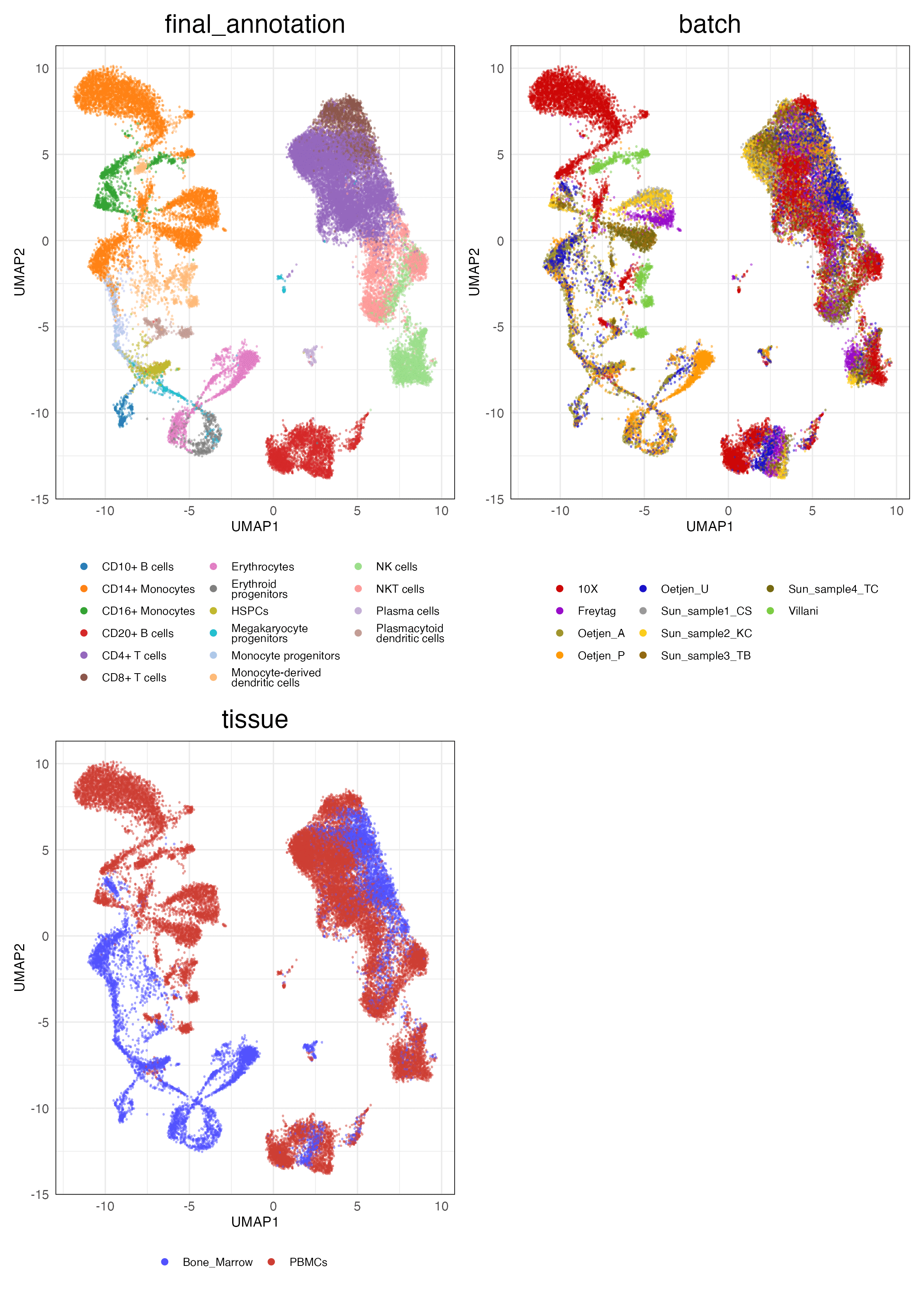
lung_atlas (Features)
Input features (Scaling)
Full (Scaled)
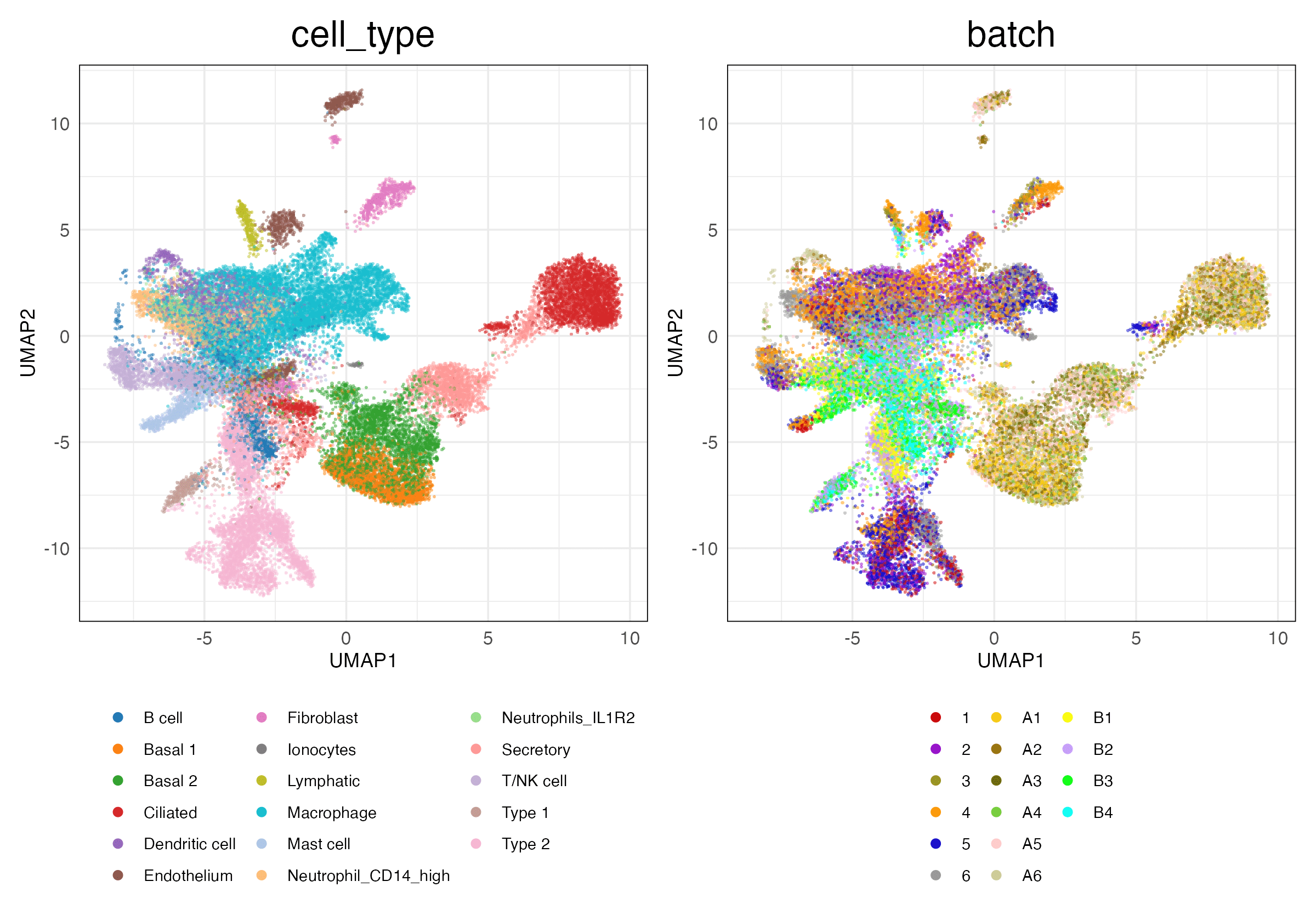
Full (Unscaled)
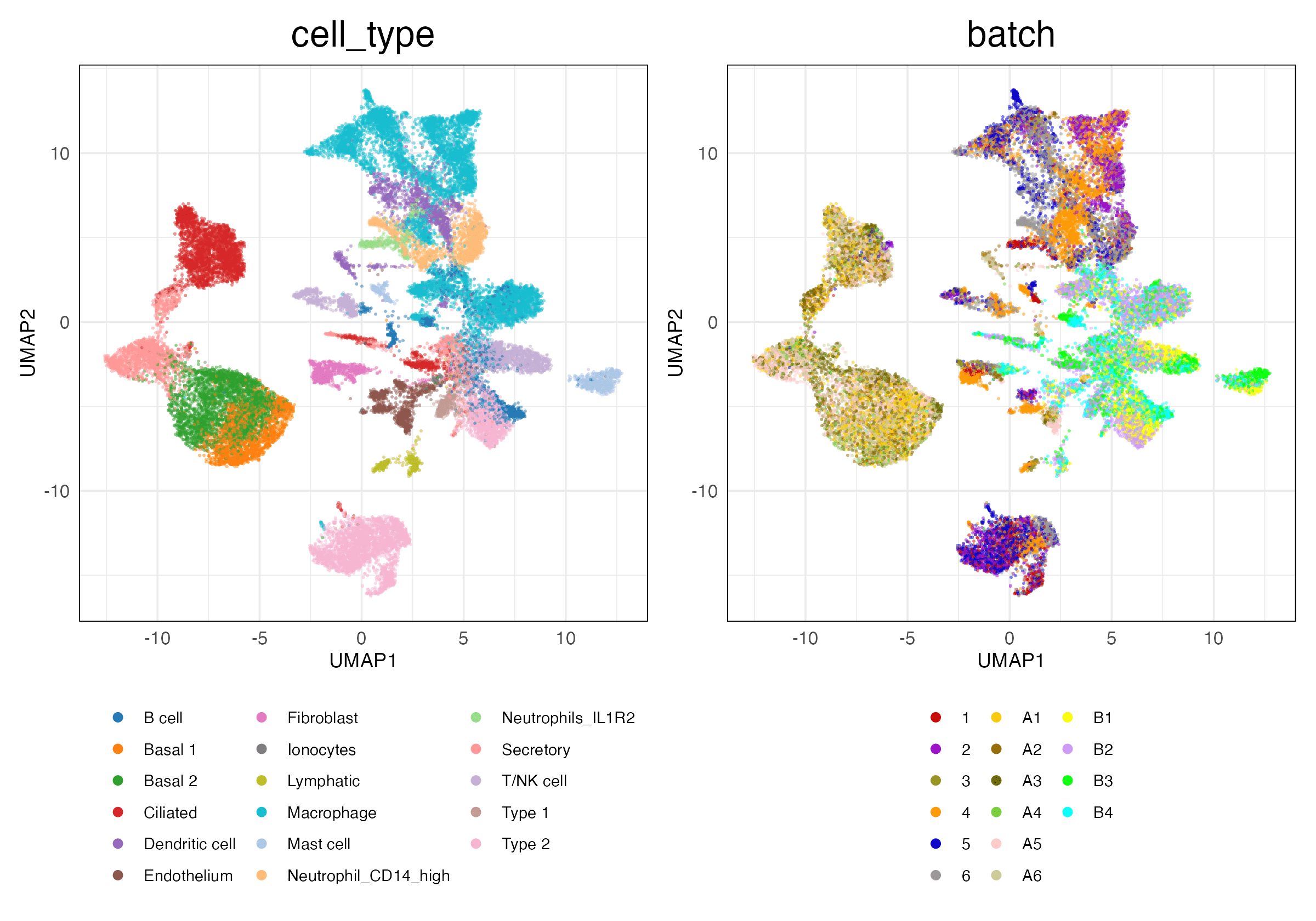
HVG (Scaled)
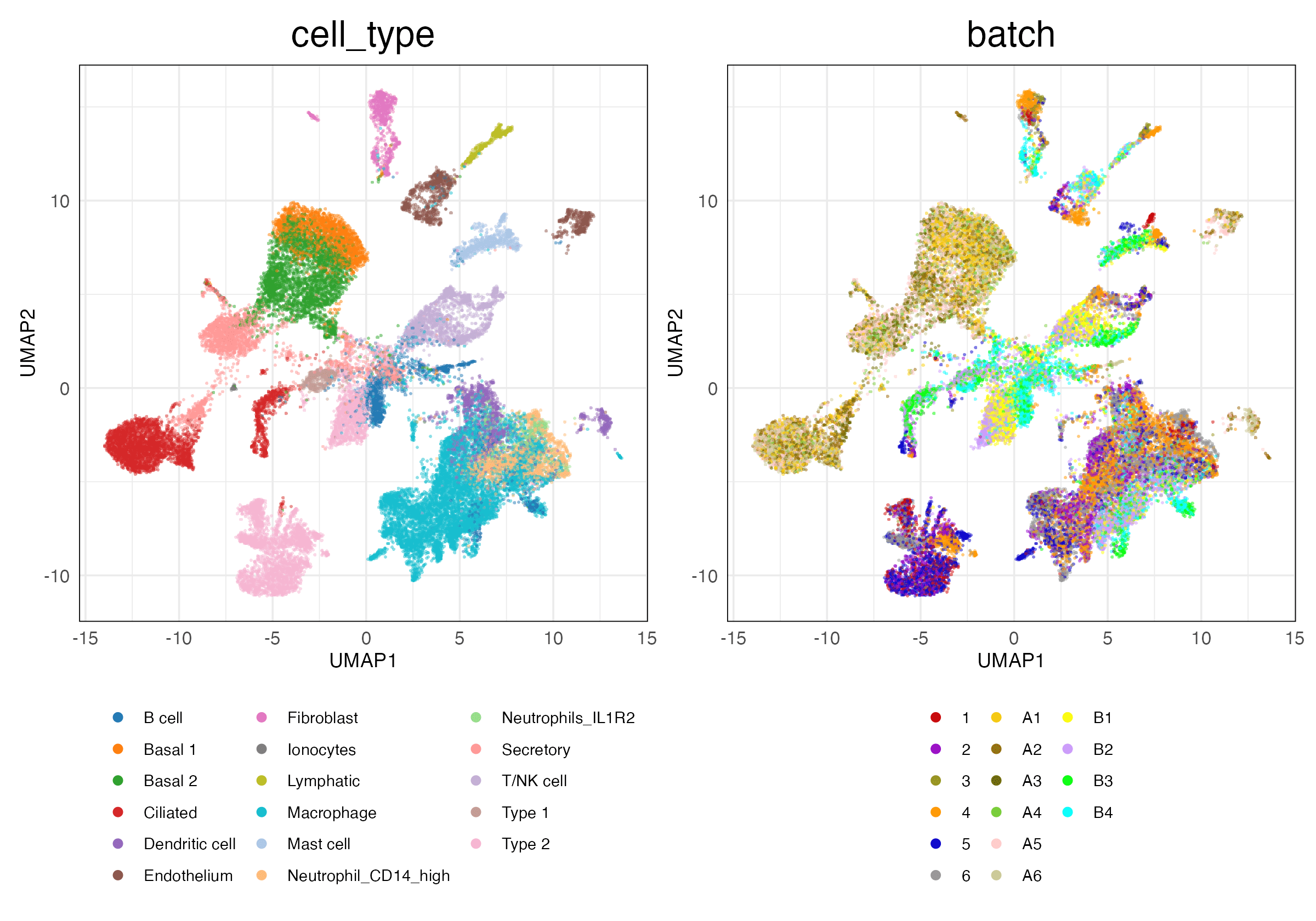
HVG (Unscaled)
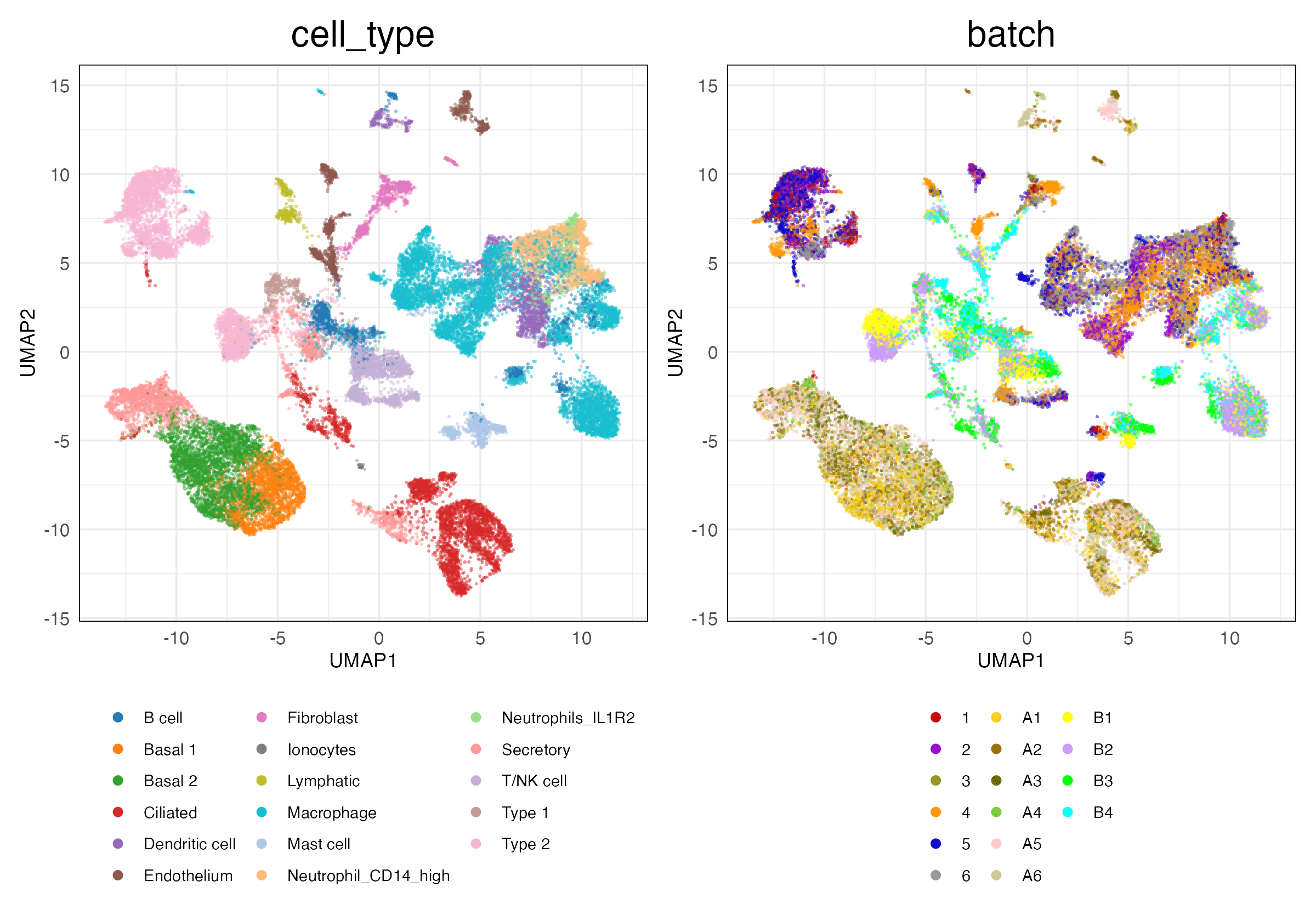
pancreas (Features)
Input features (Scaling)
Full (Scaled)
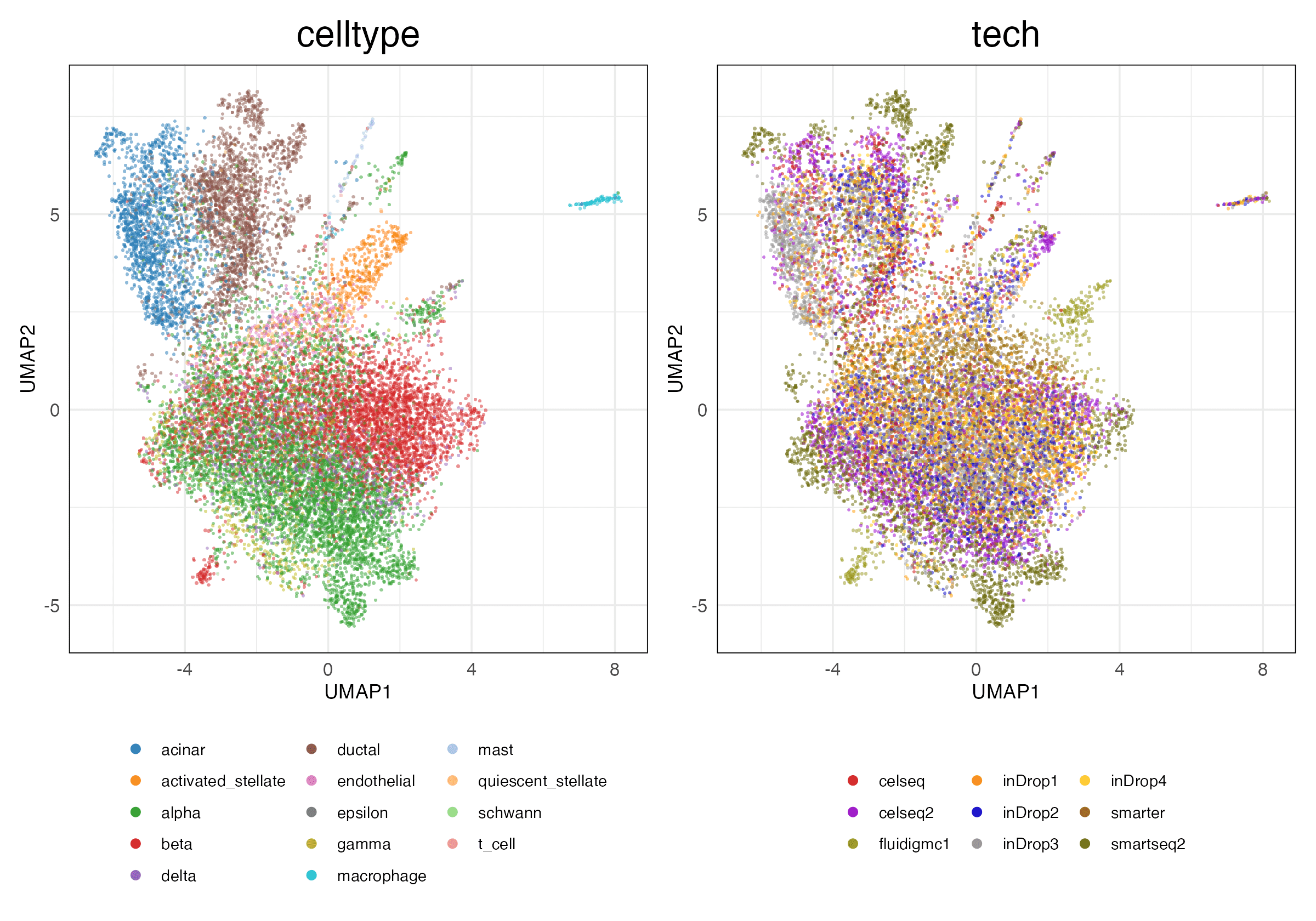
Full (Unscaled)
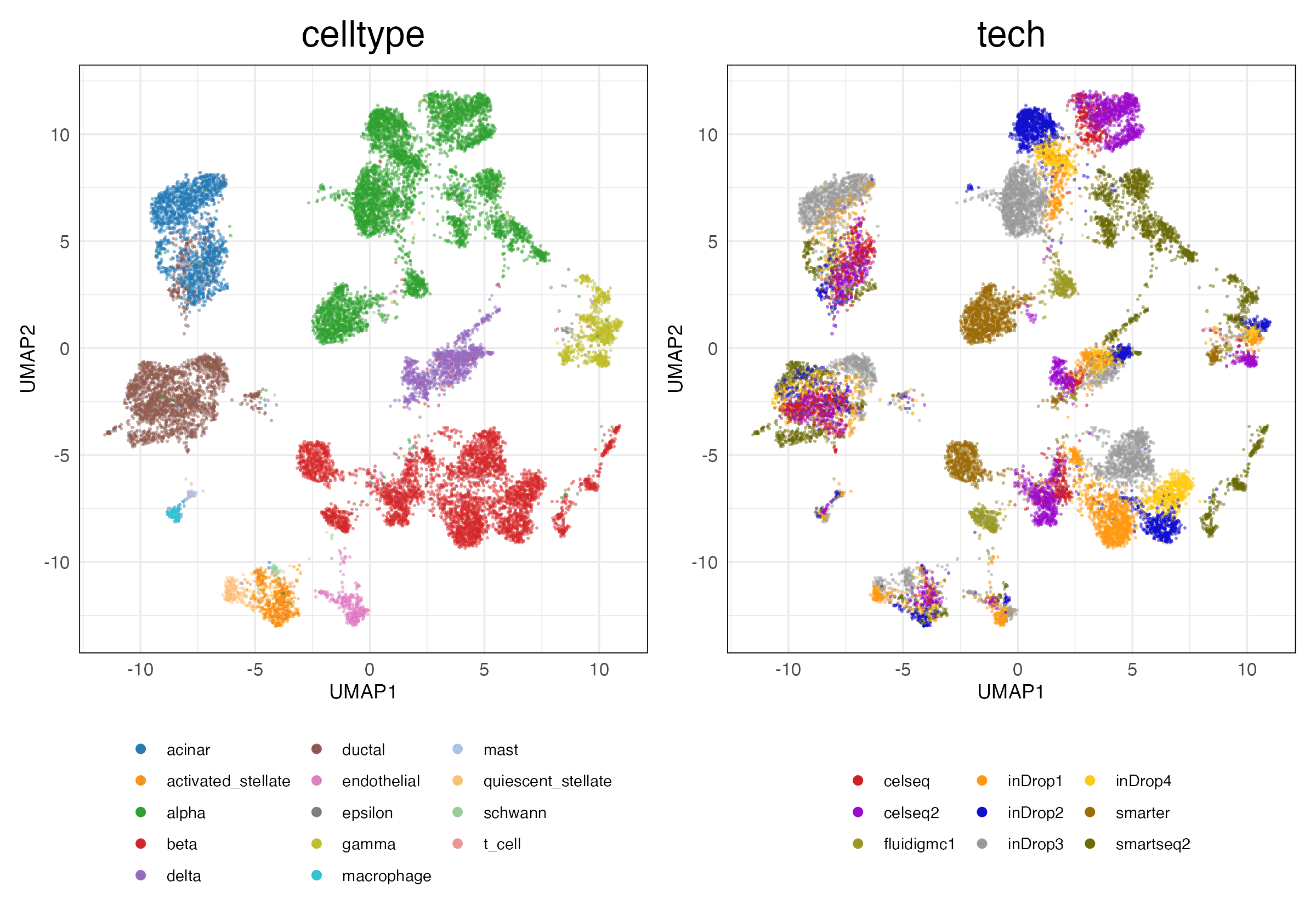
HVG (Scaled)
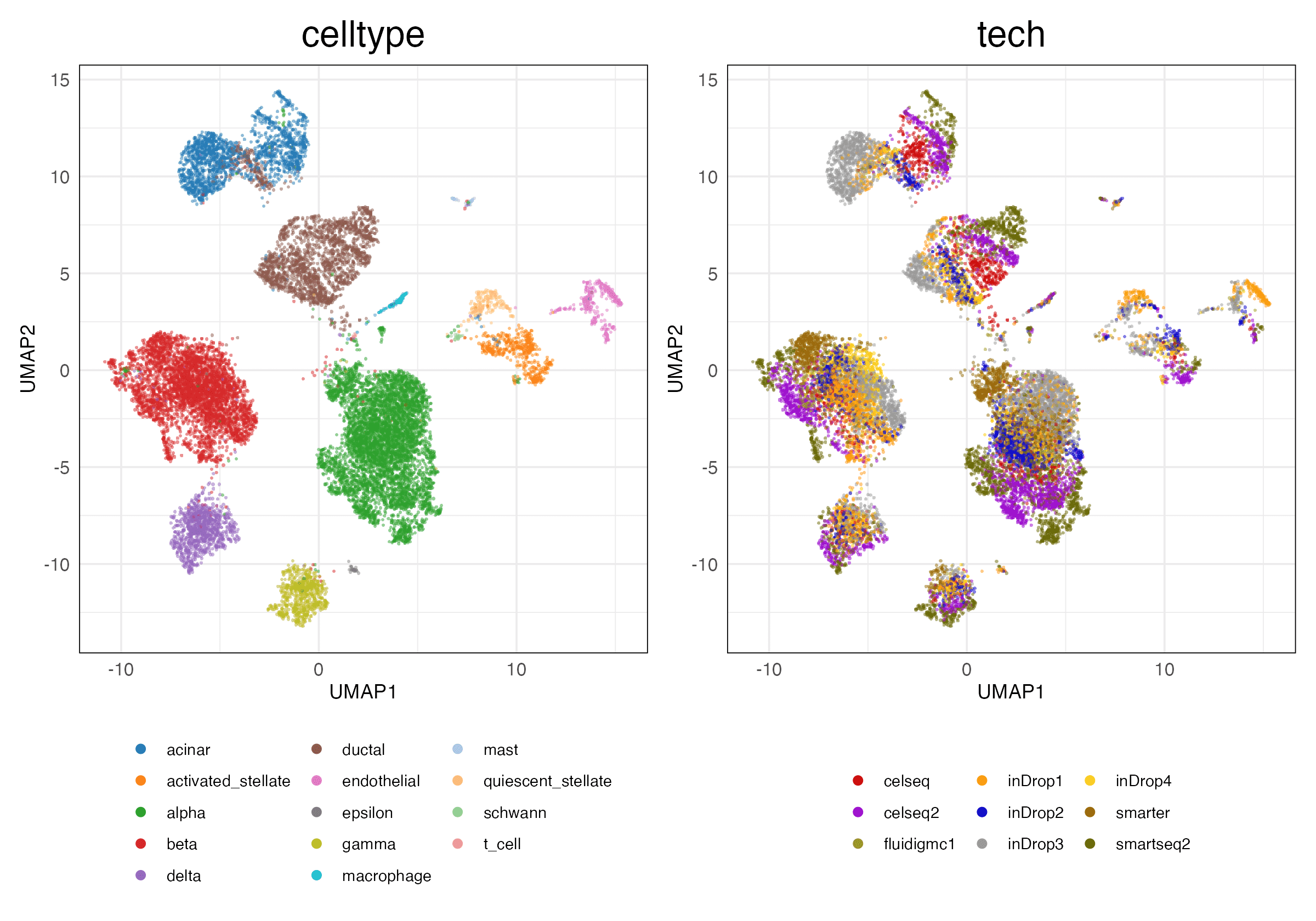
HVG (Unscaled)
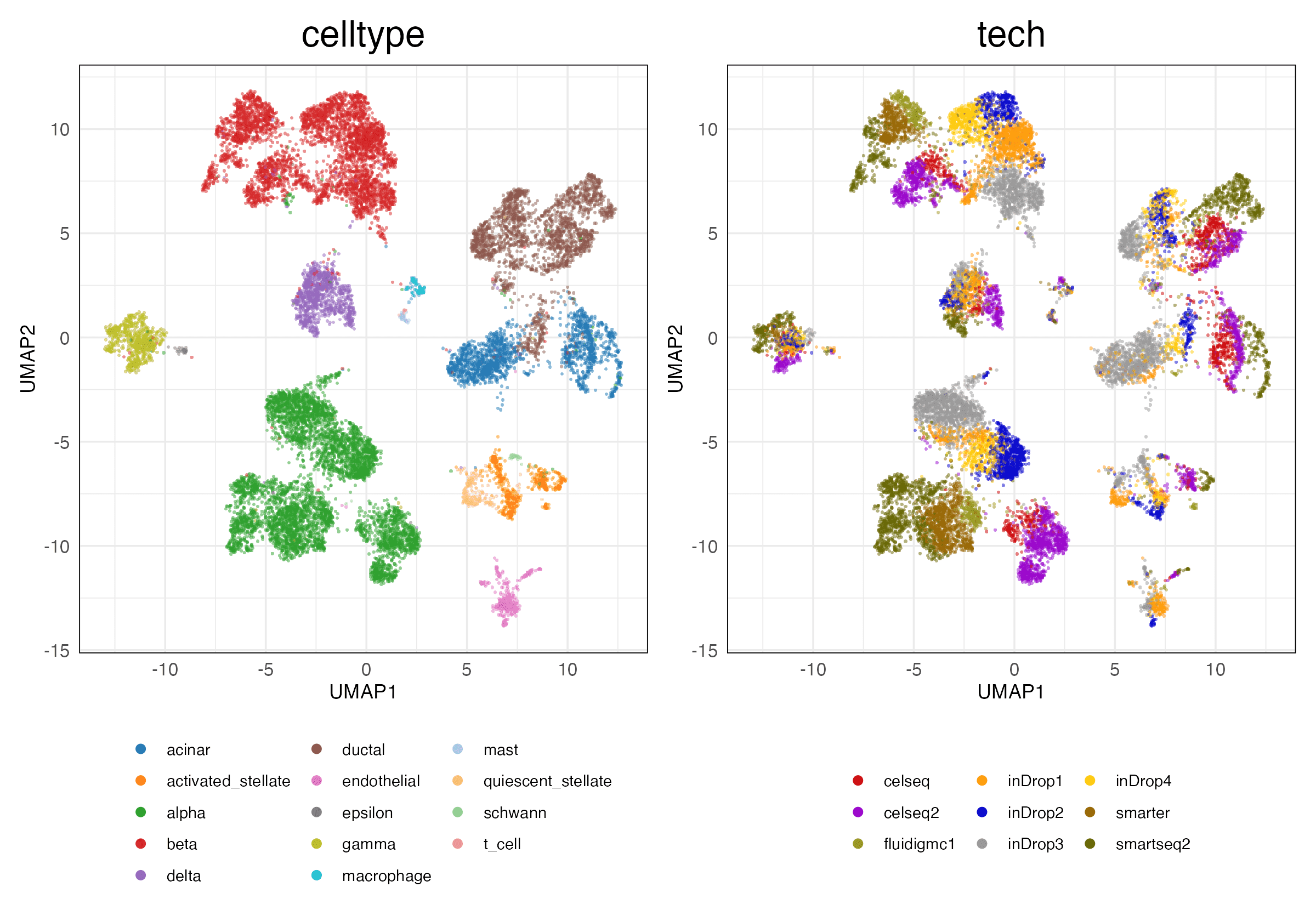
simulations_2 (Features)
Input features (Scaling)
Full (Scaled)
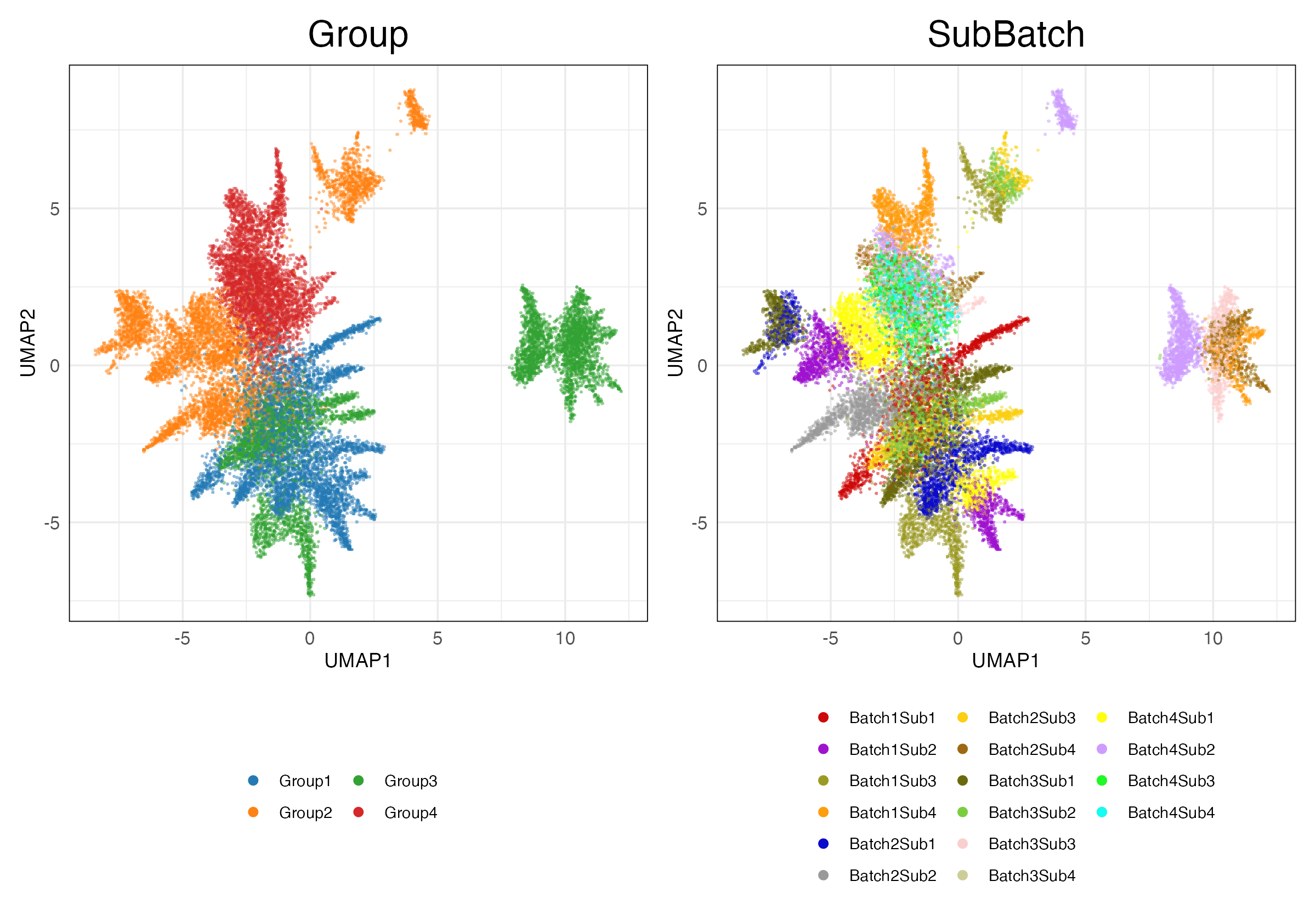
Full (Unscaled)
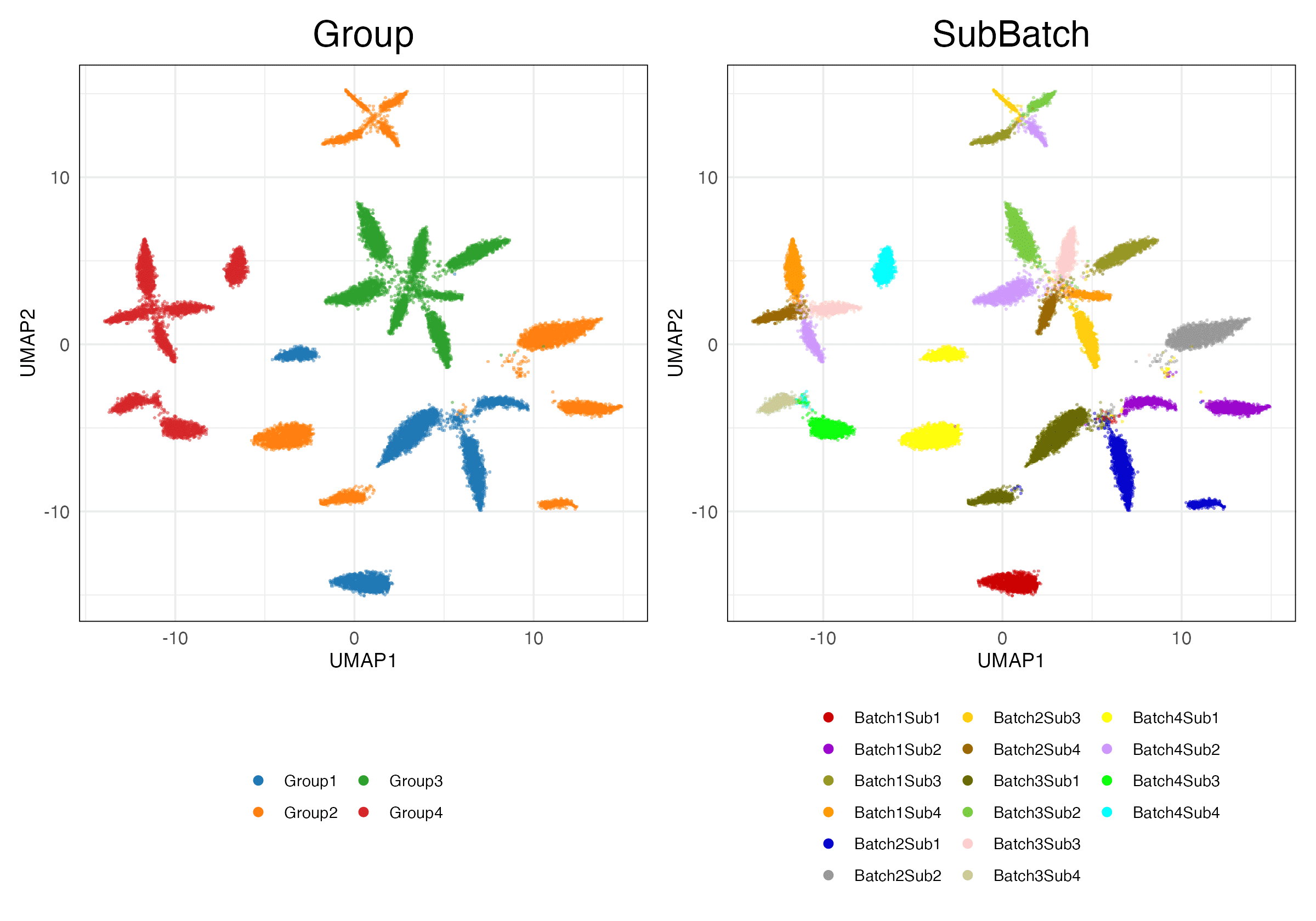
HVG (Scaled)
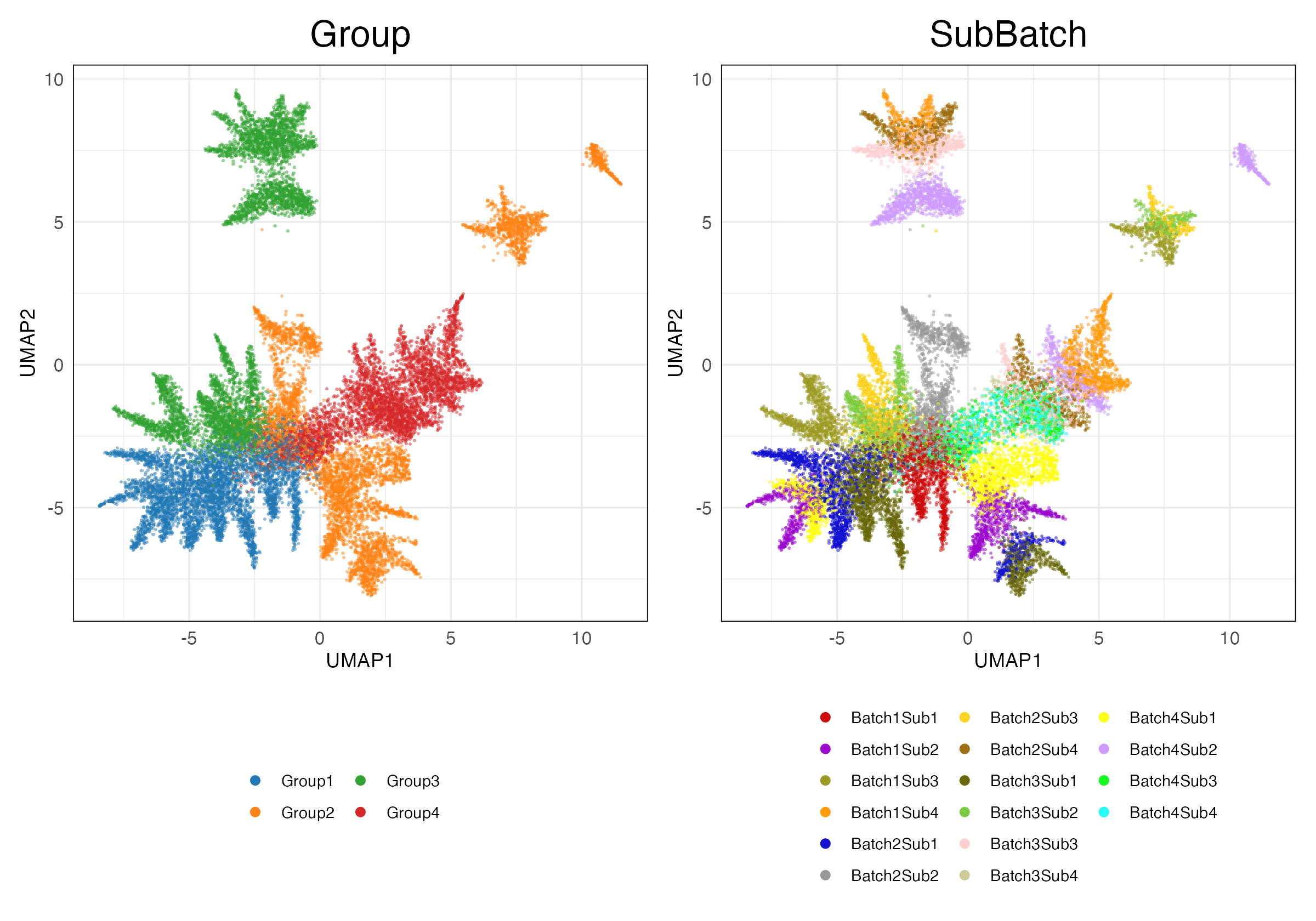
HVG (Unscaled)
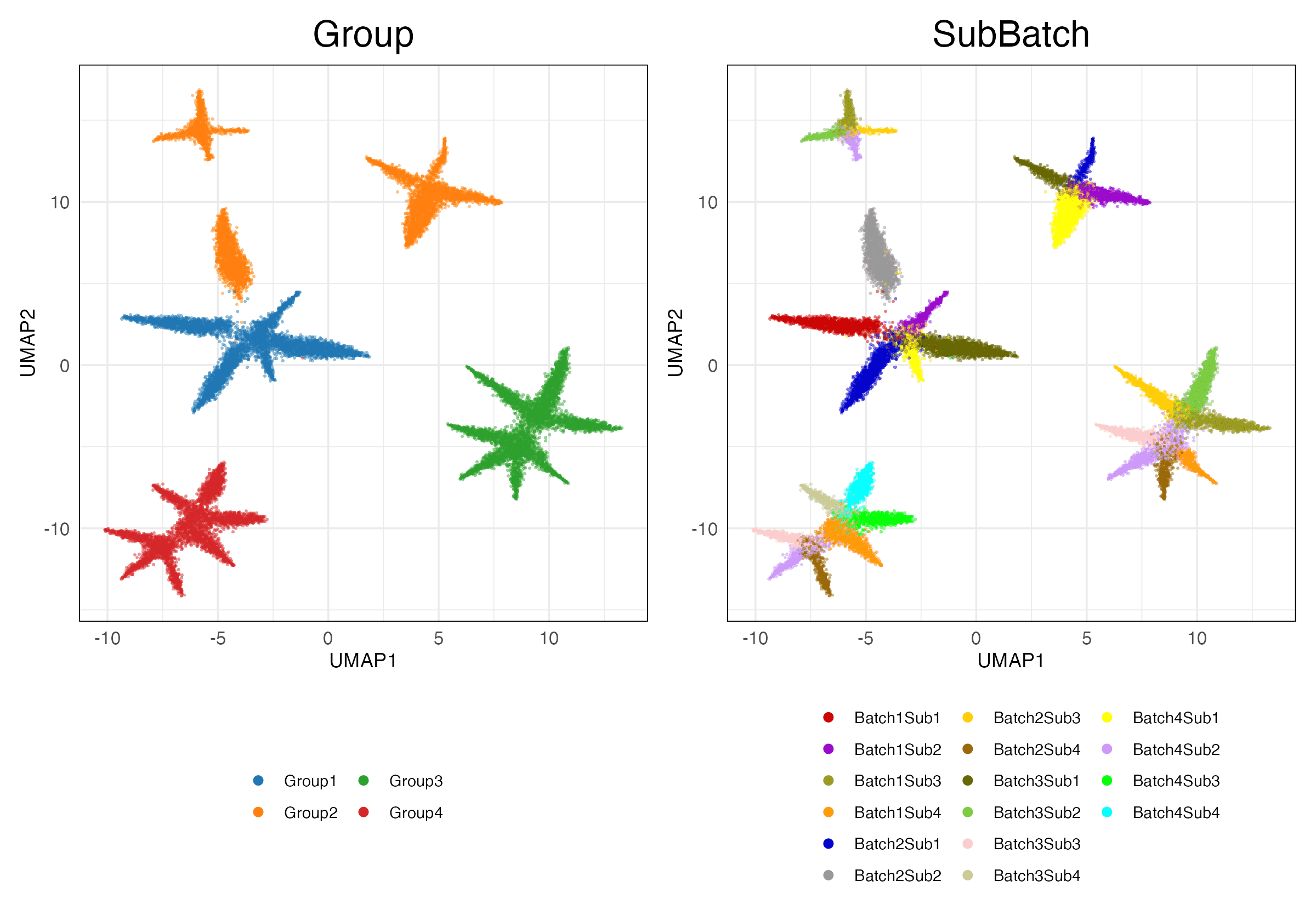
immune_cell_hum_mou (Features)
Input features (Scaling)
Full (Scaled)
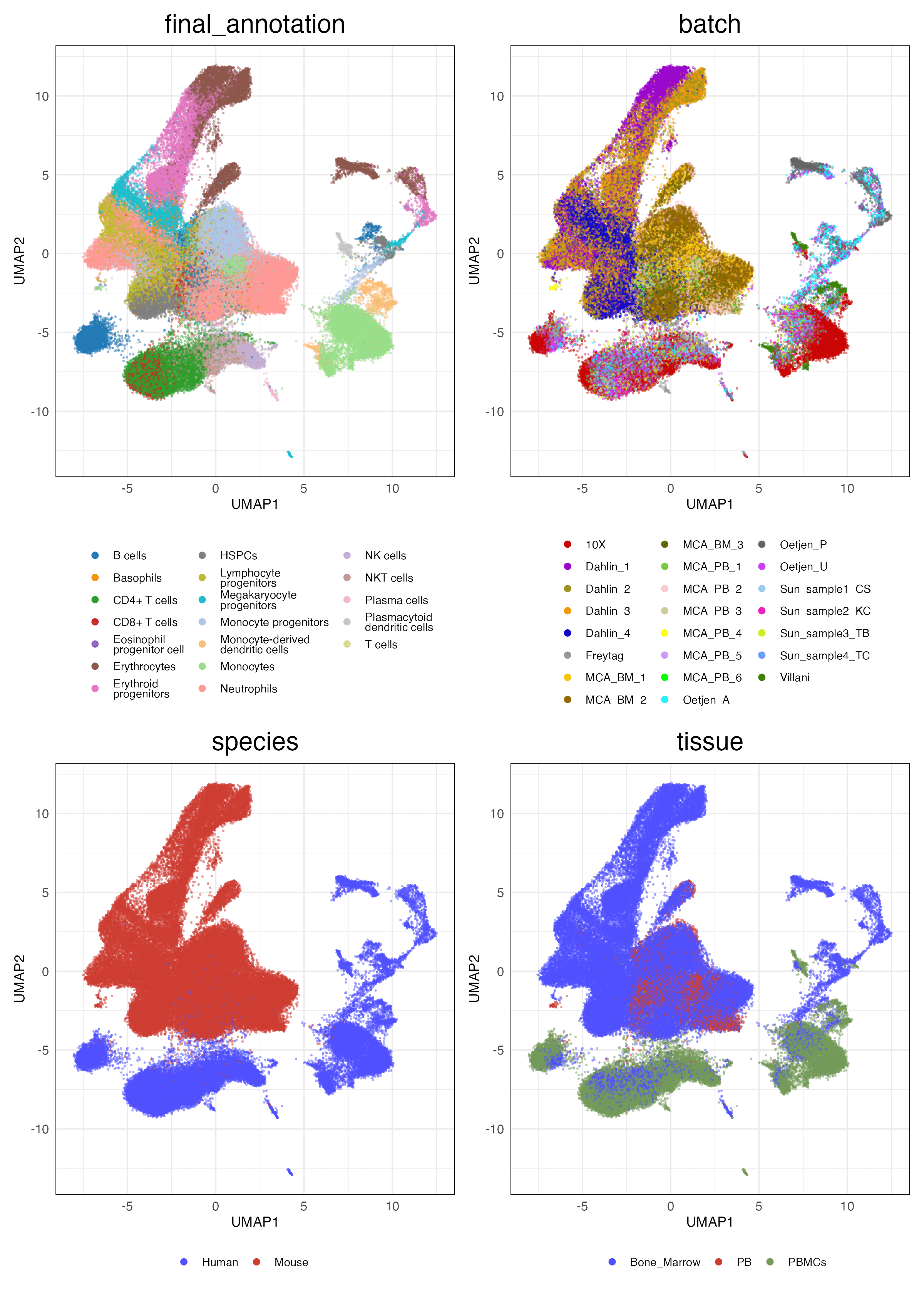
Full (Unscaled)
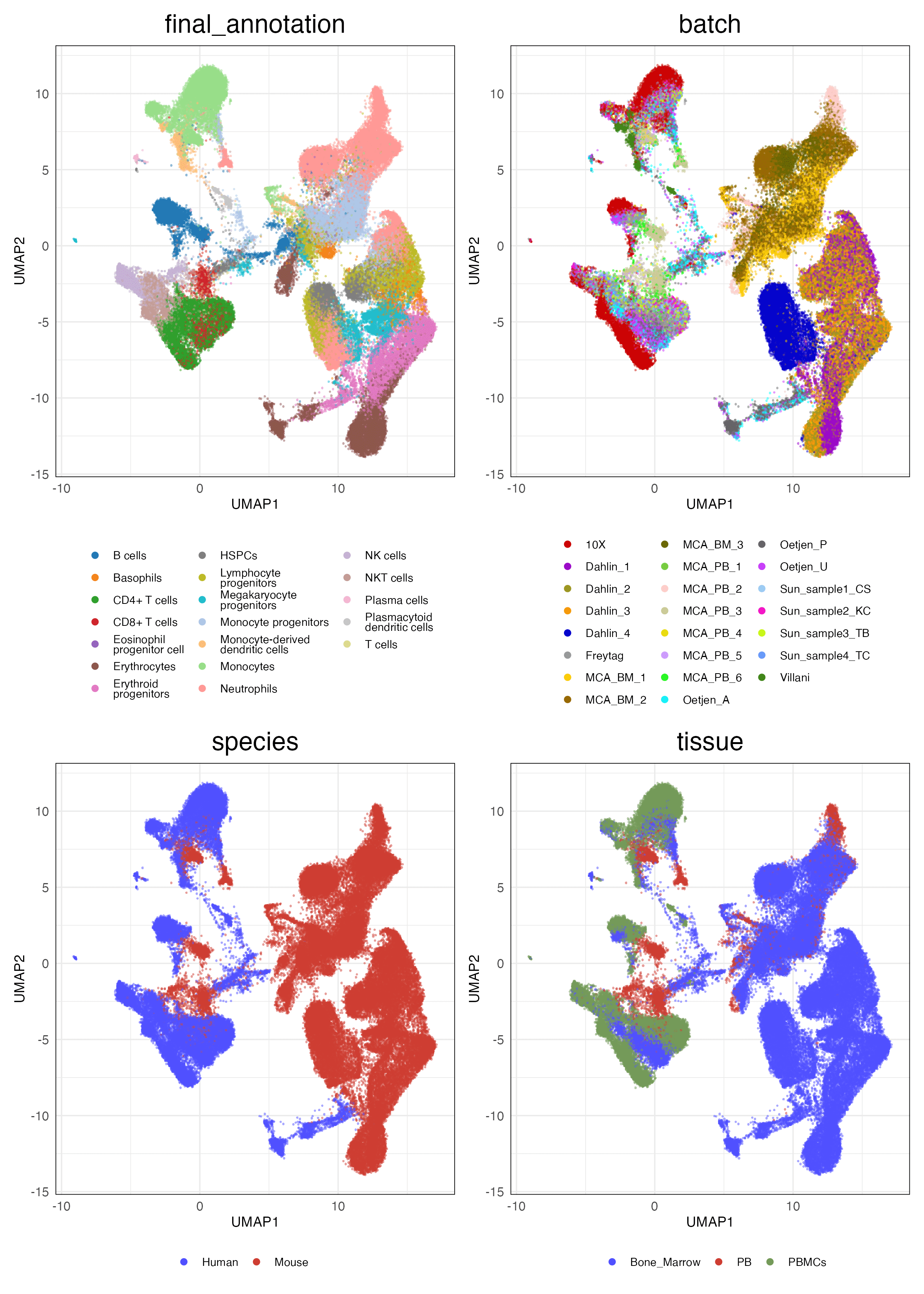
HVG (Scaled)
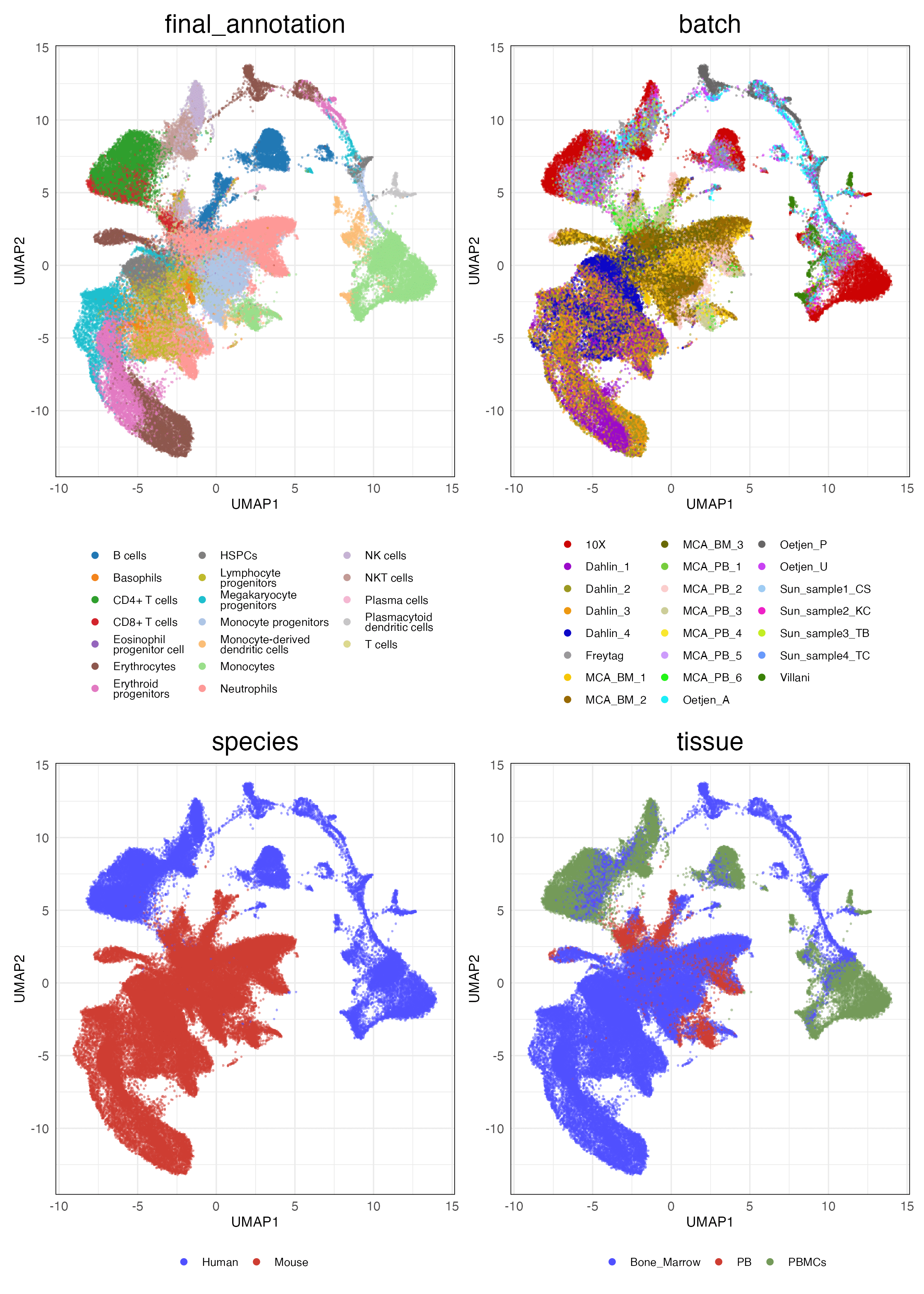
HVG (Unscaled)
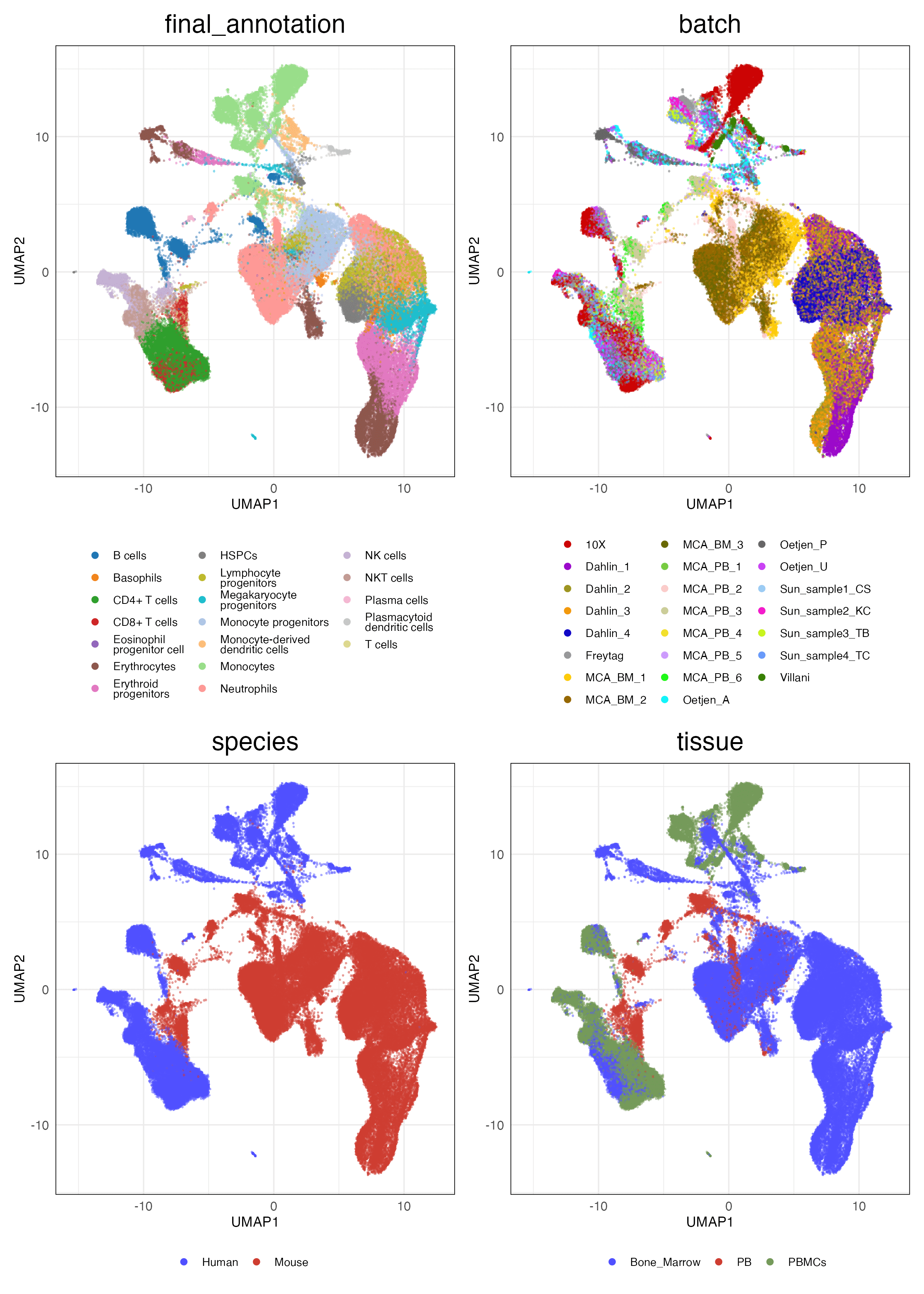
4 Scalability
4.1 Time
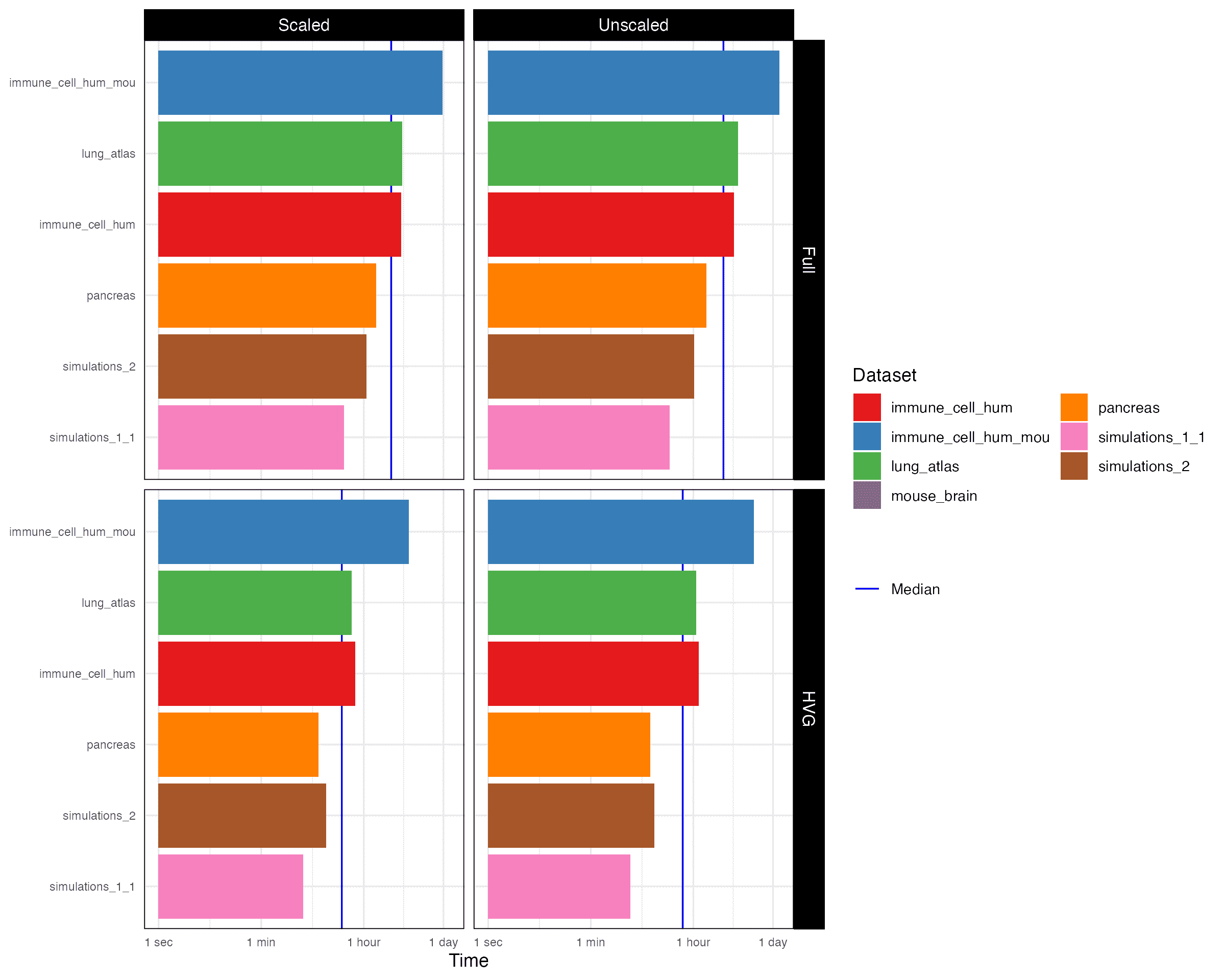
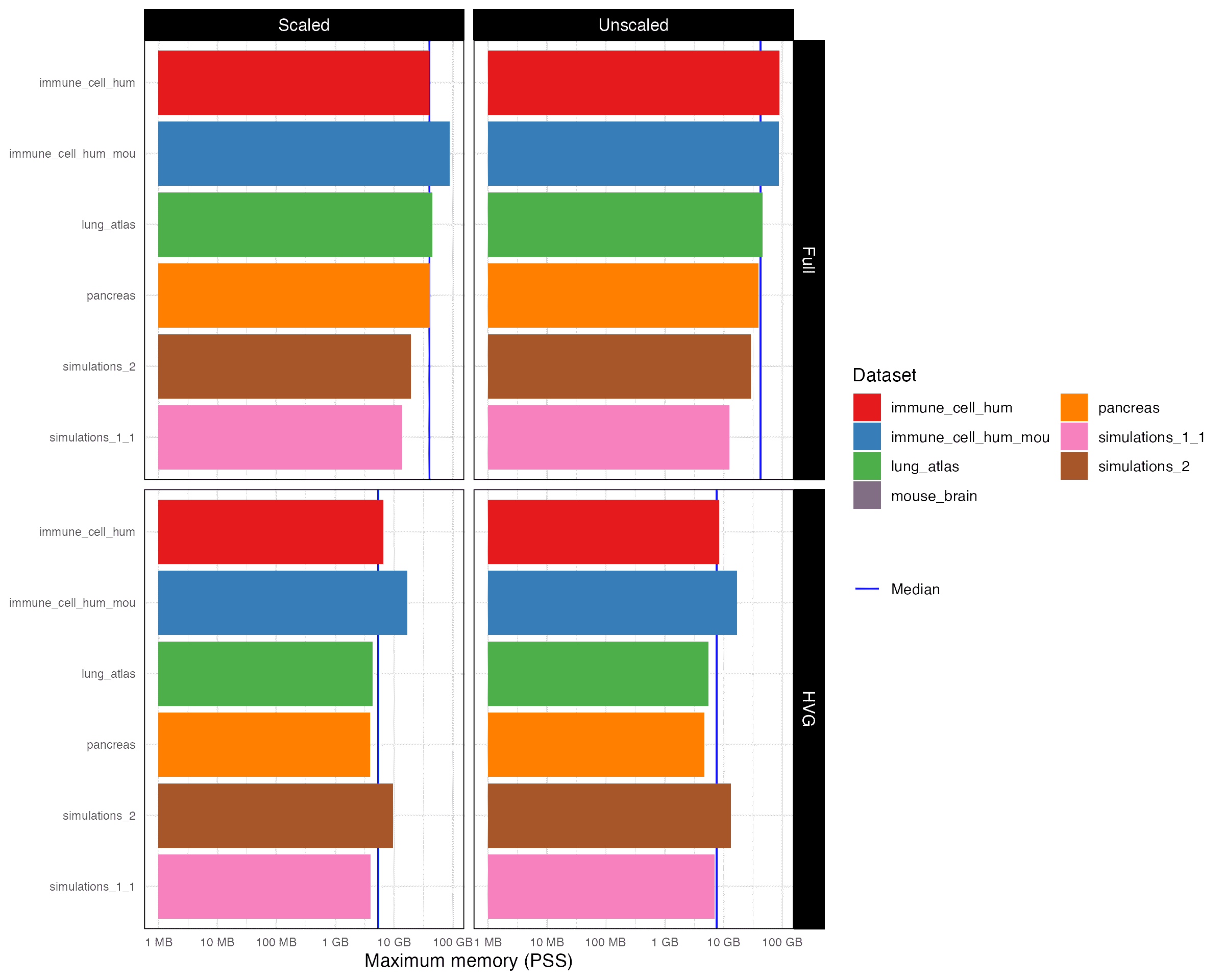
4.2 Memory