Out-of-core Analysis#
With the ever-growing size of datasets, being able to operate on larger-than-memory data is every important. Both scanpy (CPU) and rapids_singlecell (GPU) support full out-of-core, parallelizable analysis steps. The ad.experimental.read_elem_as_dask enables users to lazily load numeric matrix elements of the AnnData, such as X or obsm["X_pca"]. In this notebook, we’ll generate a PCA on an 100,000,000 cells from various cell lines with different treatments before visualizing a subset of the data. Downstream tasks could include anything from deep-learning to a linear classifier.
For individual tutorials, see here for rapids-singlecell and here for scanpy
from pathlib import Path
import numpy as np
import dask.distributed as dd
import scanpy as sc
import anndata as ad
import h5py
import dask
from collections import Counter
import pandas as pd
from tqdm import tqdm
import dask
import time
from dask.distributed import Client, LocalCluster
import dask
sc.logging.print_header()
/home/icb/weixu.wang/miniconda3/envs/rapids_singlecell/lib/python3.12/site-packages/session_info2/__init__.py:124: UserWarning: The '__version__' attribute is deprecated and will be removed in MarkupSafe 3.1. Use feature detection, or `importlib.metadata.version("markupsafe")`, instead.
and (v := getattr(pkg, "__version__", None))
/tmp/ipykernel_632503/582836007.py:19: RuntimeWarning: Failed to import dependencies for application/vnd.jupyter.widget-view+json representation. (ModuleNotFoundError: No module named 'ipywidgets')
sc.logging.print_header()
| Package | Version |
|---|---|
| numpy | 1.26.4 |
| dask | 2024.11.2 |
| scanpy | 1.11.0rc2 |
| anndata | 0.11.0rc3 |
| h5py | 3.12.1 |
| pandas | 2.2.3 |
| tqdm | 4.67.1 |
| distributed | 2024.11.2 |
| Component | Info |
| Python | 3.12.8 | packaged by conda-forge | (main, Dec 5 2024, 14:24:40) [GCC 13.3.0] |
| OS | Linux-5.14.0-427.37.1.el9_4.x86_64-x86_64-with-glibc2.34 |
| CPU | 128 logical CPU cores, x86_64 |
| GPU | ID: 0, NVIDIA A100 80GB PCIe, Driver: 560.35.03, Memory: 81920 MiB |
| Updated | 2025-03-05 16:10 |
Dependencies
| Dependency | Version |
|---|---|
| scikit-learn | 1.5.1 |
| scipy | 1.14.1 |
| zstandard | 0.23.0 |
| Pygments | 2.19.1 |
| sparse | 0.15.5 |
| prompt_toolkit | 3.0.50 |
| wcwidth | 0.2.13 |
| torch | 2.6.0 (2.6.0+cu124) |
| setproctitle | 1.3.5 |
| typing_extensions | 4.12.2 |
| igraph | 0.11.8 |
| comm | 0.2.2 |
| crc32c | 2.7.1 |
| asciitree | 0.3.3 |
| louvain | 0.8.2 |
| ipykernel | 6.29.5 |
| zarr | 2.18.3 |
| asttokens | 3.0.0 |
| tblib | 3.0.0 |
| cycler | 0.12.1 |
| numba | 0.60.0 |
| numcodecs | 0.15.1 |
| msgpack | 1.1.0 |
| pyarrow | 17.0.0 |
| lz4 | 4.3.3 |
| jedi | 0.19.2 |
| zict | 3.0.0 |
| toolz | 1.0.0 (toolz: 1.0.0, tlz: 1.0.1) |
| jaraco.collections | 5.1.0 |
| jaraco.text | 3.12.1 |
| stack_data | 0.6.3 |
| rapids-dask-dependency | 24.12.0a0 |
| pickleshare | 0.7.5 |
| tornado | 6.4.2 |
| texttable | 1.7.0 |
| pytz | 2024.1 |
| pyparsing | 3.2.1 |
| joblib | 1.4.2 |
| jupyter_client | 8.6.3 |
| pynvml | 11.4.1 |
| omegaconf | 2.3.0 |
| leidenalg | 0.10.2 |
| cupy | 13.3.0 |
| cuda-python | 12.6.0 |
| traitlets | 5.14.3 |
| platformdirs | 4.3.6 |
| rmm | 24.12.1 (24.12.01) |
| debugpy | 1.8.12 |
| psutil | 6.1.1 |
| more-itertools | 10.6.0 |
| natsort | 8.4.0 |
| legacy-api-wrap | 1.4.1 |
| pure_eval | 0.2.3 |
| Deprecated | 1.2.18 |
| threadpoolctl | 3.5.0 |
| pyzmq | 26.2.1 |
| executing | 2.1.0 |
| PyYAML | 6.0.2 |
| colorama | 0.4.6 |
| session-info2 | 0.1.2 |
| llvmlite | 0.43.0 |
| parso | 0.8.4 |
| locket | 1.0.0 |
| packaging | 24.2 |
| fastrlock | 0.8.3 |
| wrapt | 1.17.2 |
| jupyter_core | 5.7.2 |
| jaraco.functools | 4.0.1 |
| sortedcontainers | 2.4.0 |
| setuptools | 75.8.0 |
| click | 8.1.8 |
| cytoolz | 1.0.1 |
| ipython | 8.32.0 |
| jaraco.context | 5.3.0 |
| Jinja2 | 3.1.5 |
| cloudpickle | 3.1.1 |
| decorator | 5.1.1 |
| defusedxml | 0.7.1 |
| MarkupSafe | 3.0.2 |
| kiwisolver | 1.4.8 |
| pillow | 11.1.0 |
| six | 1.17.0 |
| matplotlib | 3.10.0 |
| python-dateutil | 2.9.0.post0 |
Copyable Markdown
| Package | Version | | ----------- | --------- | | numpy | 1.26.4 | | dask | 2024.11.2 | | scanpy | 1.11.0rc2 | | anndata | 0.11.0rc3 | | h5py | 3.12.1 | | pandas | 2.2.3 | | tqdm | 4.67.1 | | distributed | 2024.11.2 | | Dependency | Version | | ---------------------- | -------------------------------- | | scikit-learn | 1.5.1 | | scipy | 1.14.1 | | zstandard | 0.23.0 | | Pygments | 2.19.1 | | sparse | 0.15.5 | | prompt_toolkit | 3.0.50 | | wcwidth | 0.2.13 | | torch | 2.6.0 (2.6.0+cu124) | | setproctitle | 1.3.5 | | typing_extensions | 4.12.2 | | igraph | 0.11.8 | | comm | 0.2.2 | | crc32c | 2.7.1 | | asciitree | 0.3.3 | | louvain | 0.8.2 | | ipykernel | 6.29.5 | | zarr | 2.18.3 | | asttokens | 3.0.0 | | tblib | 3.0.0 | | cycler | 0.12.1 | | numba | 0.60.0 | | numcodecs | 0.15.1 | | msgpack | 1.1.0 | | pyarrow | 17.0.0 | | lz4 | 4.3.3 | | jedi | 0.19.2 | | zict | 3.0.0 | | toolz | 1.0.0 (toolz: 1.0.0, tlz: 1.0.1) | | jaraco.collections | 5.1.0 | | jaraco.text | 3.12.1 | | stack_data | 0.6.3 | | rapids-dask-dependency | 24.12.0a0 | | pickleshare | 0.7.5 | | tornado | 6.4.2 | | texttable | 1.7.0 | | pytz | 2024.1 | | pyparsing | 3.2.1 | | joblib | 1.4.2 | | jupyter_client | 8.6.3 | | pynvml | 11.4.1 | | omegaconf | 2.3.0 | | leidenalg | 0.10.2 | | cupy | 13.3.0 | | cuda-python | 12.6.0 | | traitlets | 5.14.3 | | platformdirs | 4.3.6 | | rmm | 24.12.1 (24.12.01) | | debugpy | 1.8.12 | | psutil | 6.1.1 | | more-itertools | 10.6.0 | | natsort | 8.4.0 | | legacy-api-wrap | 1.4.1 | | pure_eval | 0.2.3 | | Deprecated | 1.2.18 | | threadpoolctl | 3.5.0 | | pyzmq | 26.2.1 | | executing | 2.1.0 | | PyYAML | 6.0.2 | | colorama | 0.4.6 | | session-info2 | 0.1.2 | | llvmlite | 0.43.0 | | parso | 0.8.4 | | locket | 1.0.0 | | packaging | 24.2 | | fastrlock | 0.8.3 | | wrapt | 1.17.2 | | jupyter_core | 5.7.2 | | jaraco.functools | 4.0.1 | | sortedcontainers | 2.4.0 | | setuptools | 75.8.0 | | click | 8.1.8 | | cytoolz | 1.0.1 | | ipython | 8.32.0 | | jaraco.context | 5.3.0 | | Jinja2 | 3.1.5 | | cloudpickle | 3.1.1 | | decorator | 5.1.1 | | defusedxml | 0.7.1 | | MarkupSafe | 3.0.2 | | kiwisolver | 1.4.8 | | pillow | 11.1.0 | | six | 1.17.0 | | matplotlib | 3.10.0 | | python-dateutil | 2.9.0.post0 | | Component | Info | | --------- | ----------------------------------------------------------------------------- | | Python | 3.12.8 | packaged by conda-forge | (main, Dec 5 2024, 14:24:40) [GCC 13.3.0] | | OS | Linux-5.14.0-427.37.1.el9_4.x86_64-x86_64-with-glibc2.34 | | CPU | 128 logical CPU cores, x86_64 | | GPU | ID: 0, NVIDIA A100 80GB PCIe, Driver: 560.35.03, Memory: 81920 MiB | | Updated | 2025-03-05 16:10 |
Technical Notes#
To turn on one or the other of GPU or CPU, use the flag below. If you are using a GPU, configure the gpus flag accordingly to the number of GPUs you have available. Similarly, for the CPU implementation, you can configure the number of workers. We ran this notebook using CPU with a slurm configuration like srun ... -c 32 --mem=300gb, using 16 dask workers and the below sparse chunks size while we were only able to get one GPU, although the beauty of dask is that one is enough to complete the analysis thanks to the out-of-core capabilities. You can of course use fewer CPU cores and this notebook will still be runnable, such is the beauty of truly lazy, chunked computation. Nothing is loaded into memory until it is needed, and only the amount needed to perform the local computation on a chunk of the data.
The interplay between memory and workers is delicate - too many workers and not enough memory will cause your calculation to crash, so often it is good to have a few fewer workers than you think would be best. For example, because a single GPU has somewhere in the range of 32GB, that is more than the individual 16 workers can have who split the 300GB, thereby allowing for a larger chunk size. Furthermore with rapids-memory-management, memory concerns are not so overriding on the GPU.
Caution: Out-of-core support is still quite new in both packages, so we are learning more and more daily about performance optimizations. The information here about performance thus represents our best attempt to provide advice on performance. On only one GPU, preprocessing can be quite lengthy, and therefore it is advisable to use rapids only with more than one GPU (be sure to update the gpus variable accordingly to reflect this - it should be something like '0,1,2,3', for example, for four GPUs). Otherwise, we have observed that the single-GPU PCA is faster than the multi-core PCA (~12.5 min vs. ~18 min), but the time needed to bring the data back to the CPU from the GPU for writing causes the two to be comparable i.e., PCA + writing the results has the same time between one GPU and multi-core CPU with the above specs.
use_gpu = False
if use_gpu:
import rapids_singlecell as rsc
SPARSE_CHUNK_SIZE = 1_000_000
from dask_cuda import LocalCUDACluster
import rmm
import cupy as cp
from cupyx.scipy import spx
from rmm.allocators.cupy import rmm_cupy_allocator
def set_mem():
rmm.reinitialize(managed_memory=True)
cp.cuda.set_allocator(rmm_cupy_allocator)
gpus = "0" # comma separated like "0,1,2" for 3 gpus
cluster = LocalCUDACluster(CUDA_VISIBLE_DEVICES=gpus)
dask.array.register_chunk_type(spx.csr_matrix)
else:
SPARSE_CHUNK_SIZE = 100_000
cluster = LocalCluster(n_workers=16)
client = Client(cluster)
if use_gpu:
client.run(set_mem)
mod = rsc
else:
mod = sc
client
Client
Client-7912c966-f9dc-11ef-a6b7-00001049fe80
| Connection method: Cluster object | Cluster type: distributed.LocalCluster |
| Dashboard: http://127.0.0.1:8787/status |
Cluster Info
LocalCluster
3e2f9434
| Dashboard: http://127.0.0.1:8787/status | Workers: 16 |
| Total threads: 16 | Total memory: 240.00 GiB |
| Status: running | Using processes: True |
Scheduler Info
Scheduler
Scheduler-dcc86f43-8418-4c2b-bec4-f820e7038b52
| Comm: tcp://127.0.0.1:8477 | Workers: 16 |
| Dashboard: http://127.0.0.1:8787/status | Total threads: 16 |
| Started: Just now | Total memory: 240.00 GiB |
Workers
Worker: 0
| Comm: tcp://127.0.0.1:8483 | Total threads: 1 |
| Dashboard: http://127.0.0.1:8265/status | Memory: 15.00 GiB |
| Nanny: tcp://127.0.0.1:8285 | |
| Local directory: /tmp/dask-scratch-space/worker-ft3iwcso | |
Worker: 1
| Comm: tcp://127.0.0.1:8101 | Total threads: 1 |
| Dashboard: http://127.0.0.1:8487/status | Memory: 15.00 GiB |
| Nanny: tcp://127.0.0.1:8481 | |
| Local directory: /tmp/dask-scratch-space/worker-2hvdphk9 | |
Worker: 2
| Comm: tcp://127.0.0.1:8363 | Total threads: 1 |
| Dashboard: http://127.0.0.1:8061/status | Memory: 15.00 GiB |
| Nanny: tcp://127.0.0.1:8233 | |
| Local directory: /tmp/dask-scratch-space/worker-k0dybc4h | |
Worker: 3
| Comm: tcp://127.0.0.1:8155 | Total threads: 1 |
| Dashboard: http://127.0.0.1:8485/status | Memory: 15.00 GiB |
| Nanny: tcp://127.0.0.1:8499 | |
| Local directory: /tmp/dask-scratch-space/worker-gn_0piah | |
Worker: 4
| Comm: tcp://127.0.0.1:8259 | Total threads: 1 |
| Dashboard: http://127.0.0.1:8215/status | Memory: 15.00 GiB |
| Nanny: tcp://127.0.0.1:8025 | |
| Local directory: /tmp/dask-scratch-space/worker-dwdoyzsm | |
Worker: 5
| Comm: tcp://127.0.0.1:8081 | Total threads: 1 |
| Dashboard: http://127.0.0.1:8497/status | Memory: 15.00 GiB |
| Nanny: tcp://127.0.0.1:8059 | |
| Local directory: /tmp/dask-scratch-space/worker-uqt168qj | |
Worker: 6
| Comm: tcp://127.0.0.1:8301 | Total threads: 1 |
| Dashboard: http://127.0.0.1:8231/status | Memory: 15.00 GiB |
| Nanny: tcp://127.0.0.1:8255 | |
| Local directory: /tmp/dask-scratch-space/worker-ldeqv_q_ | |
Worker: 7
| Comm: tcp://127.0.0.1:8121 | Total threads: 1 |
| Dashboard: http://127.0.0.1:8293/status | Memory: 15.00 GiB |
| Nanny: tcp://127.0.0.1:8185 | |
| Local directory: /tmp/dask-scratch-space/worker-5kv9x6bx | |
Worker: 8
| Comm: tcp://127.0.0.1:8235 | Total threads: 1 |
| Dashboard: http://127.0.0.1:8055/status | Memory: 15.00 GiB |
| Nanny: tcp://127.0.0.1:8109 | |
| Local directory: /tmp/dask-scratch-space/worker-3zbts9vu | |
Worker: 9
| Comm: tcp://127.0.0.1:8191 | Total threads: 1 |
| Dashboard: http://127.0.0.1:8267/status | Memory: 15.00 GiB |
| Nanny: tcp://127.0.0.1:8373 | |
| Local directory: /tmp/dask-scratch-space/worker-jx4zbf7k | |
Worker: 10
| Comm: tcp://127.0.0.1:8375 | Total threads: 1 |
| Dashboard: http://127.0.0.1:8445/status | Memory: 15.00 GiB |
| Nanny: tcp://127.0.0.1:8237 | |
| Local directory: /tmp/dask-scratch-space/worker-u5oh644i | |
Worker: 11
| Comm: tcp://127.0.0.1:8189 | Total threads: 1 |
| Dashboard: http://127.0.0.1:8069/status | Memory: 15.00 GiB |
| Nanny: tcp://127.0.0.1:8261 | |
| Local directory: /tmp/dask-scratch-space/worker-rxe7zfzv | |
Worker: 12
| Comm: tcp://127.0.0.1:8361 | Total threads: 1 |
| Dashboard: http://127.0.0.1:8129/status | Memory: 15.00 GiB |
| Nanny: tcp://127.0.0.1:8309 | |
| Local directory: /tmp/dask-scratch-space/worker-gc99zj5u | |
Worker: 13
| Comm: tcp://127.0.0.1:8461 | Total threads: 1 |
| Dashboard: http://127.0.0.1:8357/status | Memory: 15.00 GiB |
| Nanny: tcp://127.0.0.1:8219 | |
| Local directory: /tmp/dask-scratch-space/worker-w8q0t2gp | |
Worker: 14
| Comm: tcp://127.0.0.1:8433 | Total threads: 1 |
| Dashboard: http://127.0.0.1:8263/status | Memory: 15.00 GiB |
| Nanny: tcp://127.0.0.1:8257 | |
| Local directory: /tmp/dask-scratch-space/worker-1la7lk5m | |
Worker: 15
| Comm: tcp://127.0.0.1:8243 | Total threads: 1 |
| Dashboard: http://127.0.0.1:8307/status | Memory: 15.00 GiB |
| Nanny: tcp://127.0.0.1:8067 | |
| Local directory: /tmp/dask-scratch-space/worker-k9rna9hc | |
Preprocessing#
First, let’s do some standard preprocessing on the datasets. Note that we use ad.experimental.read_elem_as_dask to load the big data, the cell x gene matrix, lazily. The preprocessing is then done lazily i.e., normalization, log1p, and highly variable gene selection. Individually, we’ll do this and then merge all the datasets into one large dataset to be used in the next step. We’ll finally merge the data into one large AnnData for the PCA. Writing this out and then reading it back in is faster than rechunking a virtually concatenated i.e., anndata.concat dataset (dask requires uniform chunks, which would not be possible without rechunking).
%%time
adatas = []
all_highly_variable_genes = []
for i in tqdm(range(14)):
id = str(i + 1)
PATH = f"/lustre/groups/ml01/workspace/weixu.wang/100m/plate{id}_filtered.h5ad"
with h5py.File(PATH, "r") as f:
adata = ad.AnnData(
obs=ad.io.read_elem(f["obs"]),
var=ad.io.read_elem(f["var"]),
)
adata.X = ad.experimental.read_elem_as_dask(
f["X"], chunks=(SPARSE_CHUNK_SIZE, adata.shape[1])
)
if use_gpu:
rsc.get.anndata_to_GPU(adata)
# 100m filtering
pass_filter_mask = adata.obs["pass_filter"] == "full"
adata = adata[pass_filter_mask, :].copy()
sc.pp.normalize_total(adata)
sc.pp.log1p(adata)
sc.pp.highly_variable_genes(adata, n_top_genes=2000)
highly_variable_genes = set(adata.var_names[adata.var["highly_variable"]])
all_highly_variable_genes.append(highly_variable_genes)
adatas.append(adata)
## select the genes appears more than two plates
gene_counts = Counter(gene for genes in all_highly_variable_genes for gene in genes)
selected_genes = {gene for gene, count in gene_counts.items() if count > 2}
for i in tqdm(range(14)):
id = str(i + 1)
common_genes = list(set(adata.var_names) & selected_genes)
adata = adatas[i][:, common_genes].copy()
output_path = f"/lustre/groups/ml01/workspace/ilan.gold/100m/plate{id}_filtered_preprocessed_{'gpu' if use_gpu else 'cpu'}.h5ad"
adata.write_h5ad(output_path)
Merge all data into giant file#
We’ll now merge the data into one large file for the PCA. This is faster than rechunking a virtually concatenated i.e., anndata.concat dataset (dask requires uniform chunks, which would not be possible without rechunking). Furthermore, rechunking can cause a memory blow-up.
%%time
data_dict = {}
for i in range(14):
data_dict.update({f"plate_{i+1}": f"/lustre/groups/ml01/workspace/ilan.gold/100m/plate{i+1}_filtered_preprocessed_{'gpu' if use_gpu else 'cpu'}.h5ad"})
ad.experimental.concat_on_disk(
data_dict,
f'/lustre/groups/ml01/workspace/ilan.gold/100m/plate_merged_{"gpu" if use_gpu else "cpu"}.h5ad',
label='plate',
)
Calculate cell embeddings#
To do this we will use PCA. Both scanpy and rapids_singlecell provide out-of-core implementations of PCA on sparse datasets.
%%time
with h5py.File(f"/lustre/groups/ml01/workspace/ilan.gold/100m/plate_merged_{'gpu' if use_gpu else 'cpu'}.h5ad", "r") as f:
adata = ad.AnnData(
obs=ad.io.read_elem(f["obs"]),
var=ad.io.read_elem(f["var"]),
)
adata.X = ad.experimental.read_elem_as_dask(
f["X"], chunks=(SPARSE_CHUNK_SIZE, adata.shape[1])
)
CPU times: user 2min 13s, sys: 59.2 s, total: 3min 12s
Wall time: 3min 40s
adata
AnnData object with n_obs × n_vars = 95624334 × 2304
obs: 'sample', 'species', 'gene_count', 'tscp_count', 'mread_count', 'bc1_wind', 'bc2_wind', 'bc3_wind', 'bc1_well', 'bc2_well', 'bc3_well', 'id', 'drugname_drugconc', 'drug', 'INT_ID', 'NUM.SNPS', 'NUM.READS', 'demuxlet_call', 'BEST.GUESS', 'BEST.LLK', 'NEXT.GUESS', 'NEXT.LLK', 'DIFF.LLK.BEST.NEXT', 'BEST.POSTERIOR', 'SNG.POSTERIOR', 'cell_line', 'SNG.BEST.LLK', 'SNG.NEXT.GUESS', 'SNG.NEXT.LLK', 'SNG.ONLY.POSTERIOR', 'DBL.BEST.GUESS', 'DBL.BEST.LLK', 'DIFF.LLK.SNG.DBL', 'sublibrary', 'BARCODE', 'pcnt_mito', 'S_score', 'G2M_score', 'phase', 'cell_line_orig', 'pass_filter', 'cell_name', 'plate'
obsm: 'X_pca'
%%time
if use_gpu:
rsc.get.anndata_to_GPU(adata)
CPU times: user 3 μs, sys: 1e+03 ns, total: 4 μs
Wall time: 29.3 μs
%%time
mod.pp.pca(adata, n_comps=300, mask_var=None)
if use_gpu:
adata.obsm["X_pca"] = adata.obsm["X_pca"].map_blocks(
lambda x: x.get(), meta=np.array([]), dtype=adata.obsm["X_pca"].dtype
) # bring back to CPU
CPU times: user 4min, sys: 26.1 s, total: 4min 26s
Wall time: 17min 11s
%%time
adata.obsm["X_pca"].to_zarr(f"/lustre/groups/ml01/workspace/ilan.gold/100m/plate_merged_pca_{'gpu' if use_gpu else 'cpu'}.zarr")
CPU times: user 2min 48s, sys: 19.3 s, total: 3min 7s
Wall time: 15min 14s
Now the data can be read back in with dask if needed and put into the anndata object. A slight more complex way to write the data, but perhaps more anndata-ic would have been to use write_elem: https://anndata.readthedocs.io/en/latest/generated/anndata.io.write_elem.html
%%time
adata.obsm["X_pca"] = dask.array.from_zarr(f"/lustre/groups/ml01/workspace/ilan.gold/100m/plate_merged_pca_{'gpu' if use_gpu else 'cpu'}.zarr")
CPU times: user 196 ms, sys: 70.1 ms, total: 266 ms
Wall time: 1.23 s
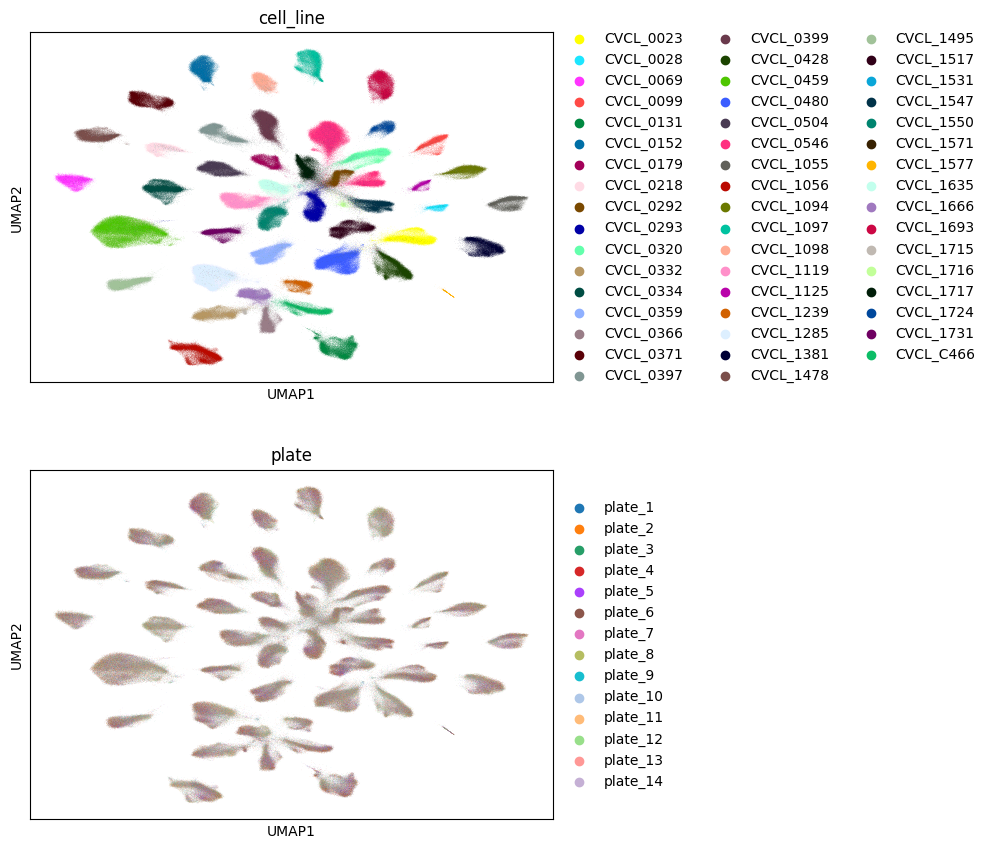
Visualization#
Briefly, we visualize a subset of the results to avoid meaningless overplotting. However, the full PCA results are still available on disk and are ready for use downstream, whether in deep-learning applications or for simple linear classifiers.
%%time
subset_adata = adata[np.random.randint(0, adata.shape[0], (1_000_000,))]
subset_adata
CPU times: user 1.56 s, sys: 676 ms, total: 2.24 s
Wall time: 3.35 s
View of AnnData object with n_obs × n_vars = 1000000 × 2304
obs: 'sample', 'species', 'gene_count', 'tscp_count', 'mread_count', 'bc1_wind', 'bc2_wind', 'bc3_wind', 'bc1_well', 'bc2_well', 'bc3_well', 'id', 'drugname_drugconc', 'drug', 'INT_ID', 'NUM.SNPS', 'NUM.READS', 'demuxlet_call', 'BEST.GUESS', 'BEST.LLK', 'NEXT.GUESS', 'NEXT.LLK', 'DIFF.LLK.BEST.NEXT', 'BEST.POSTERIOR', 'SNG.POSTERIOR', 'cell_line', 'SNG.BEST.LLK', 'SNG.NEXT.GUESS', 'SNG.NEXT.LLK', 'SNG.ONLY.POSTERIOR', 'DBL.BEST.GUESS', 'DBL.BEST.LLK', 'DIFF.LLK.SNG.DBL', 'sublibrary', 'BARCODE', 'pcnt_mito', 'S_score', 'G2M_score', 'phase', 'cell_line_orig', 'pass_filter', 'cell_name', 'plate'
obsm: 'X_pca'
%%time
if use_gpu:
subset_adata.obsm["X_pca"] = subset_adata.obsm["X_pca"].map_blocks(
spx.csr_matrix, spx.csr_matrix(np.array([])), dtype=subset_adata.obsm["X_pca"].dtype
) # bring back to GPU
else:
subset_adata.obsm["X_pca"] = subset_adata.obsm["X_pca"].compute()
/ictstr01/home/icb/ilan.gold/test_ken/venv/lib64/python3.12/site-packages/anndata/_core/index.py:173: PerformanceWarning: Slicing with an out-of-order index is generating 1044 times more chunks
return a[subset_idx]
/ictstr01/home/icb/ilan.gold/test_ken/venv/lib64/python3.12/site-packages/distributed/client.py:3357: UserWarning: Sending large graph of size 49.22 MiB.
This may cause some slowdown.
Consider scattering data ahead of time and using futures.
warnings.warn(
<timed exec>:1: ImplicitModificationWarning: Setting element `.obsm['X_pca']` of view, initializing view as actual.
/ictstr01/home/icb/ilan.gold/test_ken/venv/lib64/python3.12/site-packages/anndata/_core/anndata.py:1756: UserWarning: Observation names are not unique. To make them unique, call `.obs_names_make_unique`.
utils.warn_names_duplicates("obs")
CPU times: user 9min 20s, sys: 35.3 s, total: 9min 56s
Wall time: 10min 35s
/ictstr01/home/icb/ilan.gold/test_ken/venv/lib64/python3.12/site-packages/anndata/_core/anndata.py:1756: UserWarning: Observation names are not unique. To make them unique, call `.obs_names_make_unique`.
utils.warn_names_duplicates("obs")
%%time
mod.pp.neighbors(subset_adata)
/ictstr01/home/icb/ilan.gold/test_ken/venv/lib64/python3.12/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
CPU times: user 10min 5s, sys: 24.9 s, total: 10min 30s
Wall time: 9min 40s
%%time
mod.tl.umap(subset_adata)
if use_gpu:
subset_adata.obsm["X_umap"] = subset_adata.obsm["X_umap"].get()
CPU times: user 2h 35min 37s, sys: 25.3 s, total: 2h 36min 3s
Wall time: 23min 32s
%%time
sc.pl.umap(
subset_adata,
color=["cell_line", "plate"],
ncols=1
)
CPU times: user 11 s, sys: 514 ms, total: 11.5 s
Wall time: 14.7 s