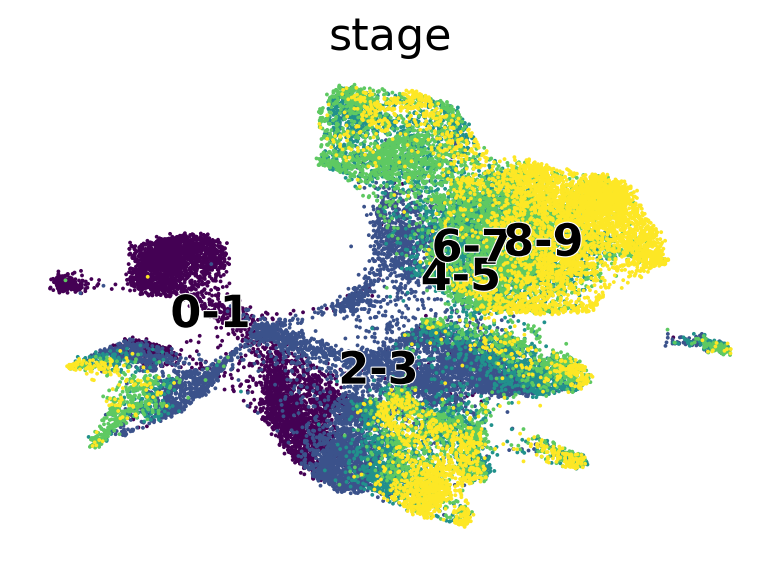
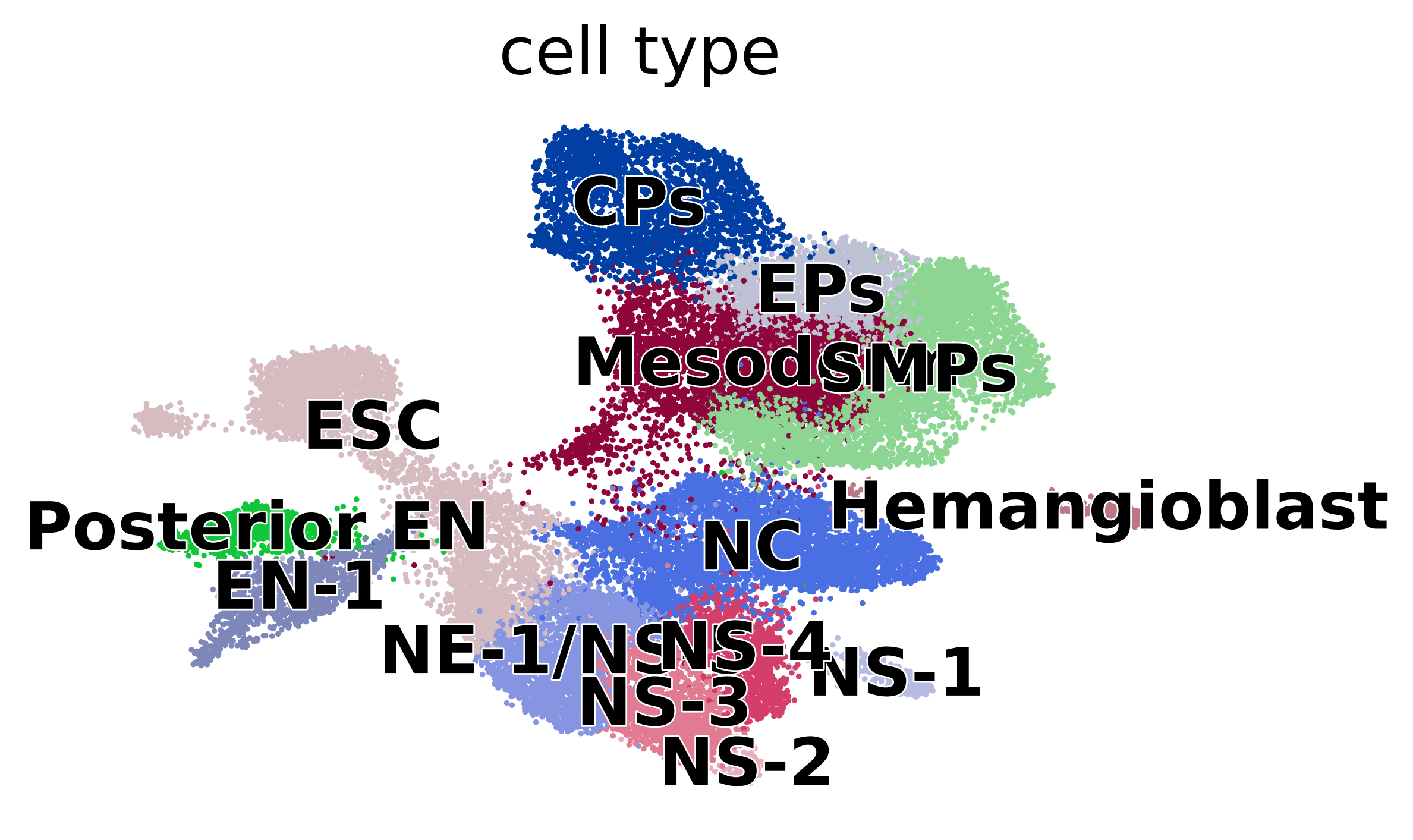
Diffusion pseudotime#
Library imports#
import sys
import matplotlib.pyplot as plt
import seaborn as sns
import scanpy as sc
import scvelo as scv
sys.path.extend(["../../../", "."])
from paths import DATA_DIR, FIG_DIR # isort: skip # noqa: E402
Global seed set to 0
General settings#
sc.settings.verbosity = 2
scv.settings.verbosity = 3
scv.settings.set_figure_params("scvelo", dpi_save=400, dpi=80, transparent=True, fontsize=20, color_map="viridis")
SAVE_FIGURES = False
if SAVE_FIGURES:
(FIG_DIR / "cytotrace_kernel" / "embryoid_body").mkdir(parents=True, exist_ok=True)
FIGURE_FORMAT = "pdf"
Data loading#
adata = sc.read(DATA_DIR / "embryoid_body" / "embryoid_body.h5ad")
adata
AnnData object with n_obs × n_vars = 31029 × 19122
obs: 'stage', 'n_genes_by_counts', 'total_counts', 'total_counts_mt', 'pct_counts_mt', 'leiden', 'cell_type'
var: 'n_cells', 'mt', 'n_cells_by_counts', 'mean_counts', 'pct_dropout_by_counts', 'total_counts', 'highly_variable', 'means', 'dispersions', 'dispersions_norm'
uns: 'hvg', 'leiden', 'log1p', 'neighbors', 'pca', 'umap'
obsm: 'X_pca', 'X_umap'
varm: 'PCs'
obsp: 'connectivities', 'distances'
Pseudotime construction#
sc.tl.diffmap(adata)
computing Diffusion Maps using n_comps=15(=n_dcs)
computing transitions
finished (0:00:00)
eigenvalues of transition matrix
[1. 0.99504757 0.99104744 0.98765516 0.98503 0.98268324
0.97936326 0.9733297 0.9698383 0.9653132 0.9609289 0.95480794
0.9547446 0.9493147 0.9468985 ]
finished (0:00:01)
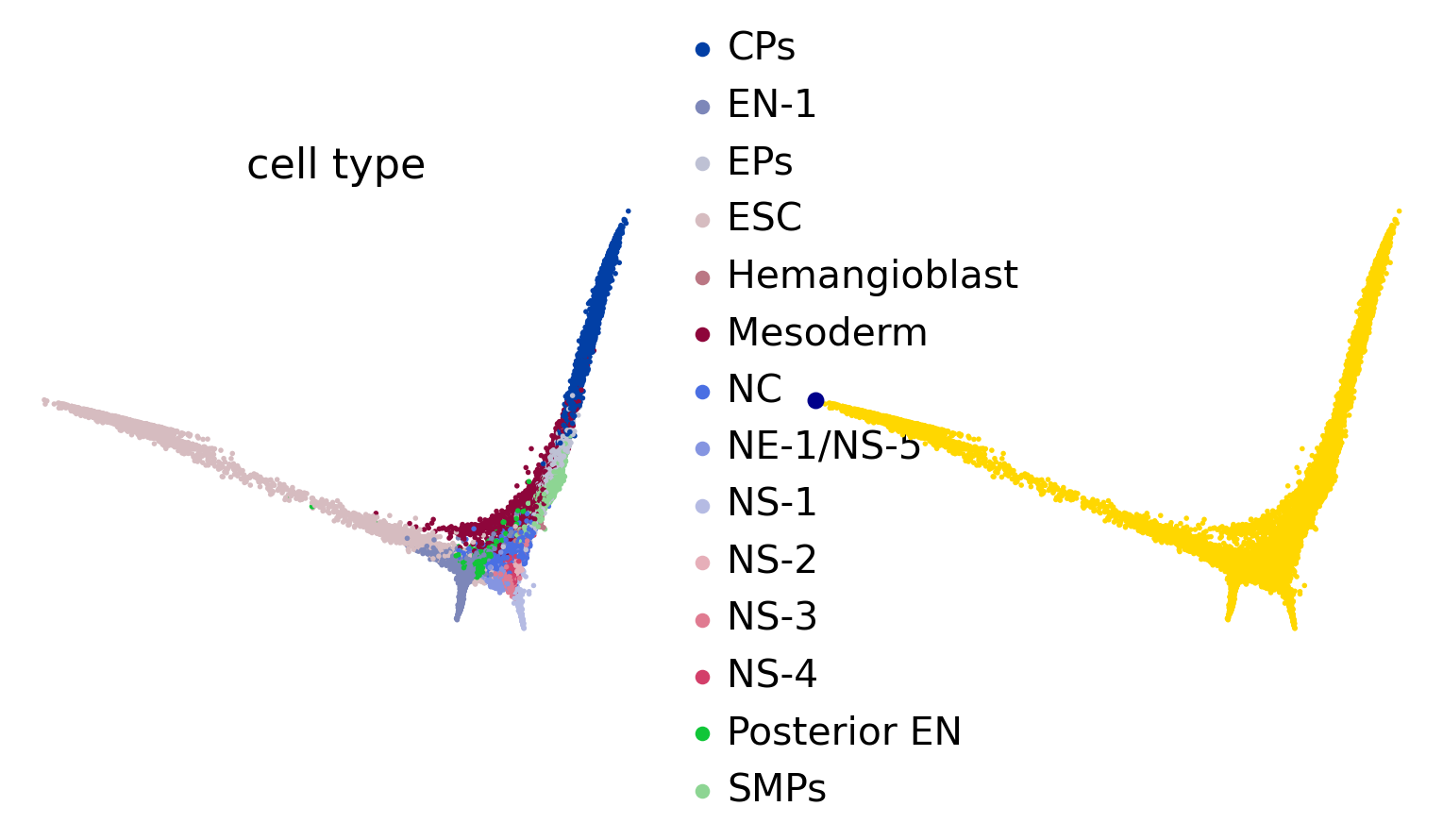
root_idx = 1458 # adata.obsm['X_diffmap'][:, 1].argmin()
scv.pl.scatter(adata, basis="diffmap", color=["cell_type", root_idx], legend_loc="right", components="1, 2", size=25)
adata.uns["iroot"] = root_idx
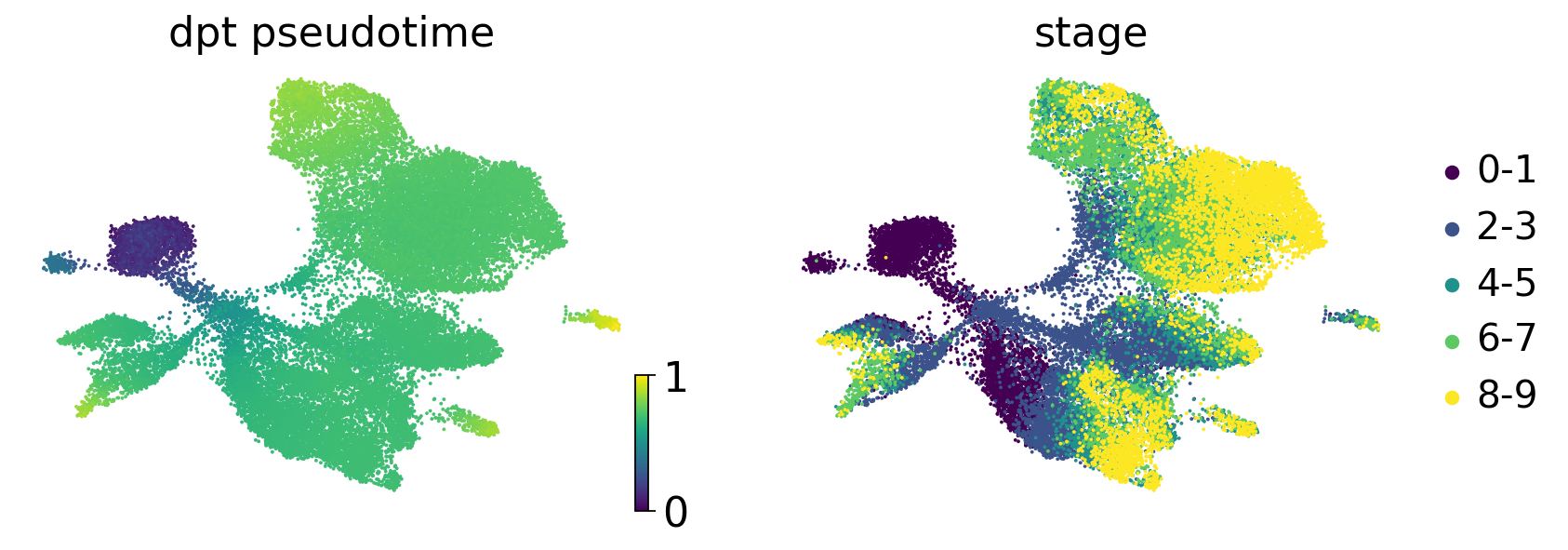
dpt_pseudotime = sc.tl.dpt(adata)
computing Diffusion Pseudotime using n_dcs=10
finished (0:00:00)
scv.pl.scatter(
adata,
c=["dpt_pseudotime", "stage"],
basis="umap",
legend_loc="right",
color_map="viridis",
)
if SAVE_FIGURES:
fig, ax = plt.subplots(figsize=(6, 4))
scv.pl.scatter(
adata, basis="umap", c="dpt_pseudotime", title="", legend_loc=False, colorbar=False, cmap="gnuplot2", ax=ax
)
fig.savefig(
FIG_DIR / "cytotrace_kernel" / "embryoid_body" / f"umap_colored_by_dpt_pseudotime.{FIGURE_FORMAT}",
format=FIGURE_FORMAT,
transparent=True,
bbox_inches="tight",
)
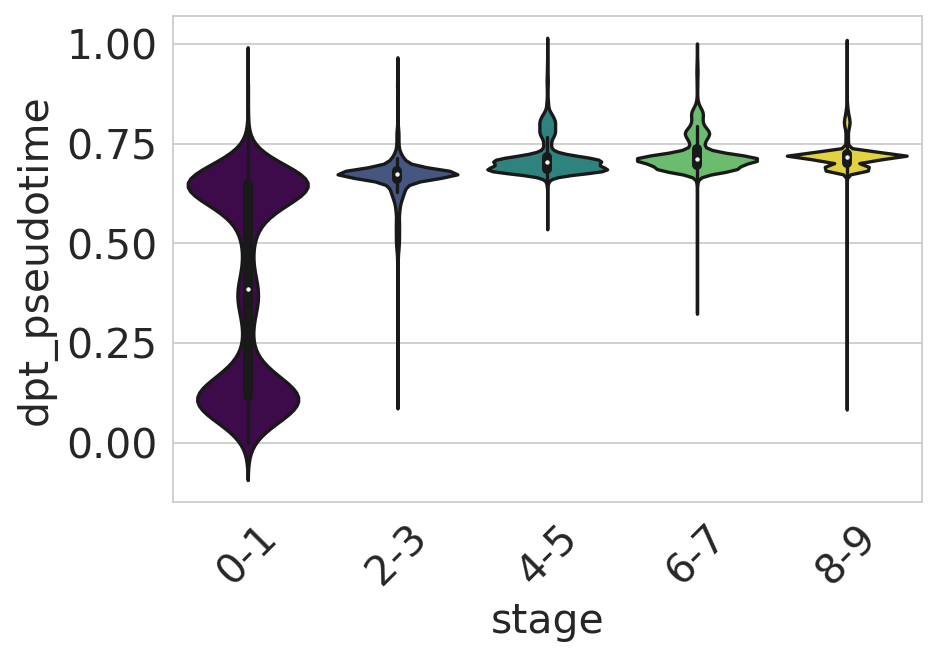
df = adata.obs[["dpt_pseudotime", "stage"]].copy()
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(6, 4))
sns.violinplot(
data=df,
x="stage",
y="dpt_pseudotime",
scale="width",
palette=["#440154", "#3b528b", "#21918c", "#5ec962", "#fde725"],
ax=ax,
)
ax.tick_params(axis="x", rotation=45)
ax.set_yticks([0, 0.25, 0.5, 0.75, 1])
plt.show()
if SAVE_FIGURES:
ax.set(xlabel=None, xticklabels=[], ylabel=None, yticklabels=[])
fig.savefig(
FIG_DIR / "cytotrace_kernel" / "embryoid_body" / f"dpt_vs_stage.{FIGURE_FORMAT}",
format=FIGURE_FORMAT,
transparent=True,
bbox_inches="tight",
)
sns.reset_orig()