RNA velocity analysis on mouse embryonic fibroblasts#
Library imports#
import sys
import pandas as pd
import matplotlib.pyplot as plt
import mplscience
import seaborn as sns
import cellrank as cr
import scanpy as sc
import scvelo as scv
from anndata import AnnData
from cr2 import running_in_notebook
sys.path.extend(["../../../", "."])
from paths import DATA_DIR, FIG_DIR # isort: skip # noqa: E402
Global seed set to 0
General settings#
# set verbosity levels
sc.settings.verbosity = 2
cr.settings.verbosity = 4
scv.settings.verbosity = 3
scv.settings.set_figure_params("scvelo", dpi_save=400, dpi=80, transparent=True, fontsize=20, color_map="viridis")
scv.settings.plot_prefix = ""
SAVE_FIGURES = False
if SAVE_FIGURES:
(FIG_DIR / "realtime_kernel" / "mef").mkdir(parents=True, exist_ok=True)
FIGURE_FORMAT = "pdf"
(DATA_DIR / "mef" / "results").mkdir(parents=True, exist_ok=True)
N_JOBS = 8
Function definitions#
Data loading#
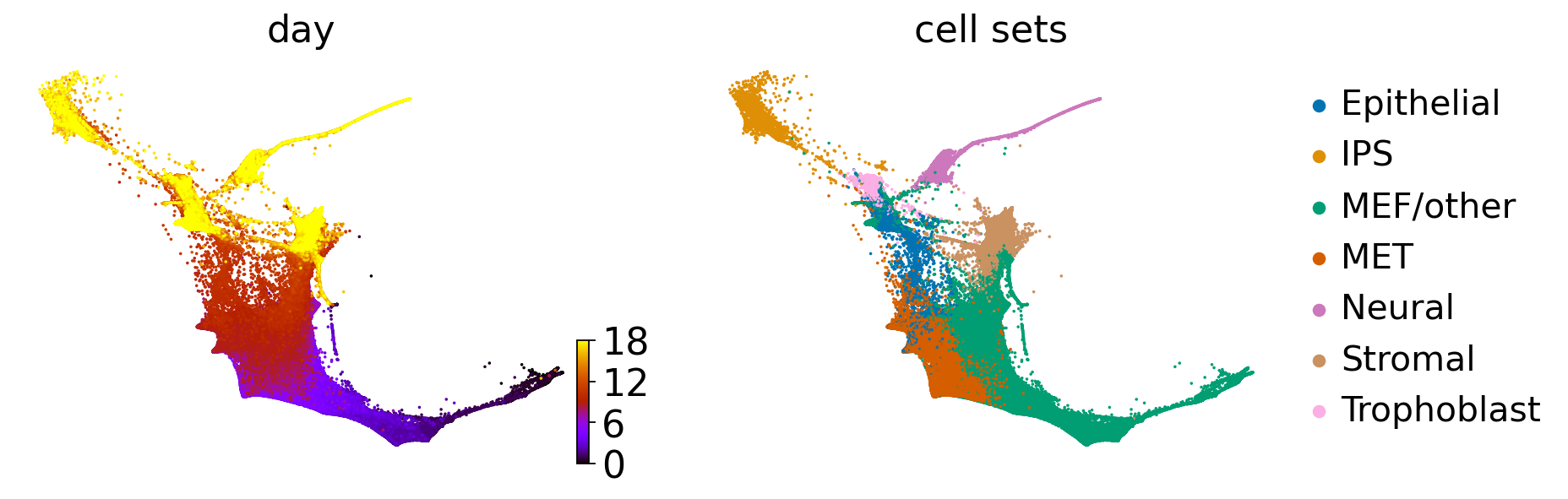
adata = sc.read(DATA_DIR / "mef" / "processed" / "mef_velo.h5ad")
adata
AnnData object with n_obs × n_vars = 236285 × 27998
obs: 'day', 'MEF.identity', 'Pluripotency', 'Cell.cycle', 'ER.stress', 'Epithelial.identity', 'ECM.rearrangement', 'Apoptosis', 'SASP', 'Neural.identity', 'Placental.identity', 'X.reactivation', 'XEN', 'Trophoblast', 'Trophoblast progenitors', 'Spiral Artery Trophpblast Giant Cells', 'Spongiotrophoblasts', 'Oligodendrocyte precursor cells (OPC)', 'Astrocytes', 'Cortical Neurons', 'RadialGlia-Id3', 'RadialGlia-Gdf10', 'RadialGlia-Neurog2', 'Long-term MEFs', 'Embryonic mesenchyme', 'Cxcl12 co-expressed', 'Ifitm1 co-expressed', 'Matn4 co-expressed', '2-cell', '4-cell', '8-cell', '16-cell', '32-cell', 'cell_growth_rate', 'serum', '2i', 'major_cell_sets', 'cell_sets', 'batch'
uns: 'batch_colors', 'cell_sets_colors', 'day_colors', 'major_cell_sets_colors'
obsm: 'X_force_directed'
layers: 'ambiguous', 'matrix', 'spliced', 'unspliced'
adata = adata[adata.obs["serum"] == "True"].copy()
adata.obs["day"] = adata.obs["day"].astype(float)
adata.uns["cell_sets_colors"] = sns.color_palette("colorblind").as_hex()[: len(adata.obs["cell_sets"].cat.categories)]
Data pre-processing and RNA velocity inference#
if (DATA_DIR / "mef" / "results" / "adata_velo_fit.h5ad").is_file():
adata = sc.read(DATA_DIR / "mef" / "results" / "adata_velo_fit.h5ad")
else:
scv.pp.filter_and_normalize(adata, min_counts=20, n_top_genes=2000)
sc.pp.pca(adata)
sc.pp.neighbors(adata, random_state=0)
scv.pp.moments(adata, n_pcs=None, n_neighbors=None)
scv.tl.recover_dynamics(adata, n_jobs=N_JOBS)
scv.tl.velocity(adata, mode="dynamical")
adata.write(DATA_DIR / "mef" / "results" / "adata_velo_fit.h5ad", compression="gzip")
CellRank analysis#
Kernel#
vk = cr.kernels.VelocityKernel(adata).compute_transition_matrix()
ck = cr.kernels.ConnectivityKernel(adata).compute_transition_matrix()
combined_kernel = 0.8 * vk + 0.2 * ck
Computing transition matrix using `'deterministic'` model
Using `softmax_scale=4.4299`
Finish (0:01:29)
Computing transition matrix based on `adata.obsp['connectivities']`
DEBUG: Density normalizing the transition matrix
Finish (0:00:00)
Estimator#
estimator = cr.estimators.GPCCA(combined_kernel)
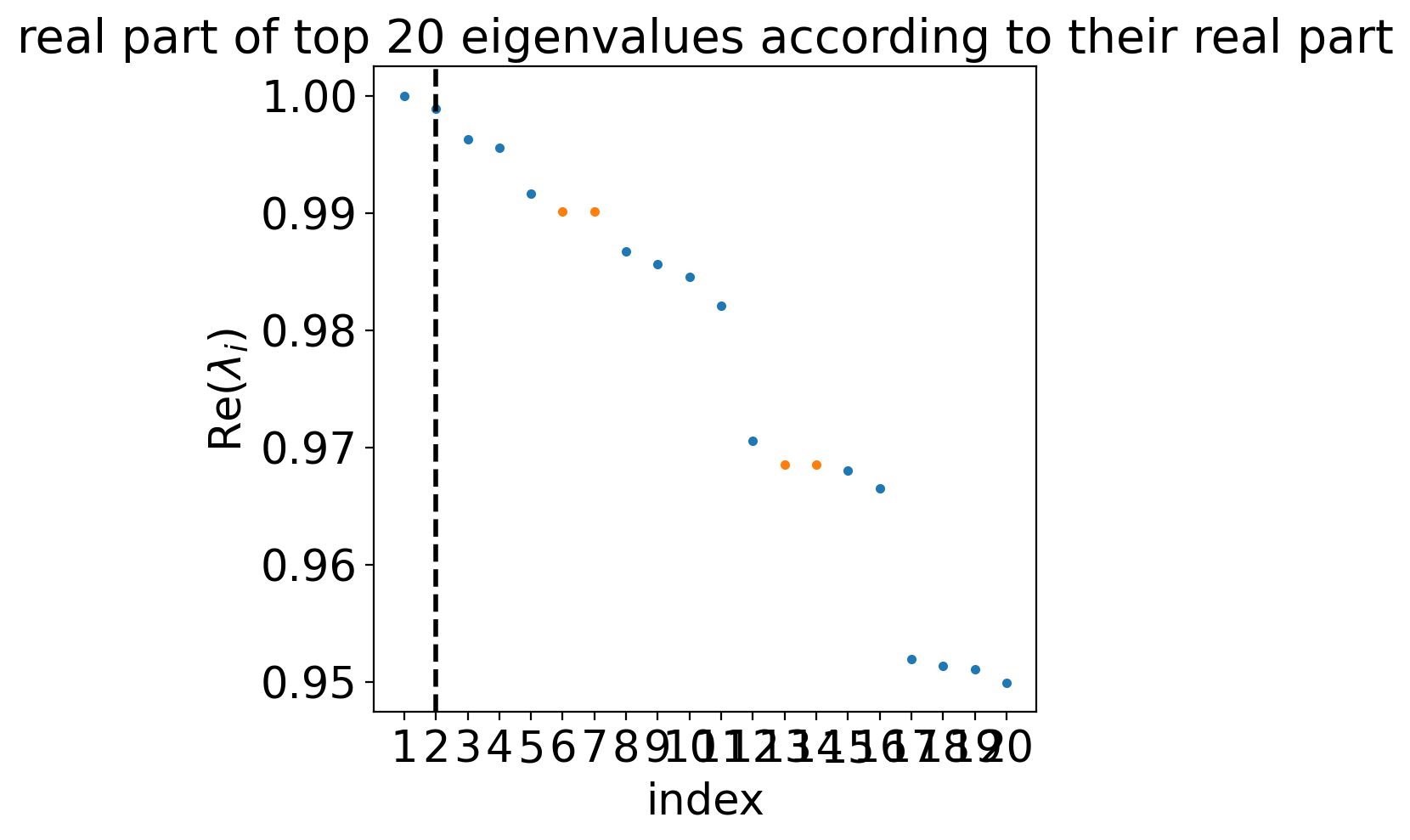
estimator.compute_schur(n_components=20)
estimator.plot_spectrum(real_only=True)
terminal_states = ["Neural", "IPS", "Trophoblast", "Stromal"]
cluster_key = "cell_sets"
if (DATA_DIR / "mef" / "results" / "tsi-vk.csv").is_file():
tsi_df = pd.read_csv(DATA_DIR / "mef" / "results" / "tsi-vk.csv")
estimator._tsi = AnnData(tsi_df, uns={"terminal_states": terminal_states, "cluster_key": cluster_key})
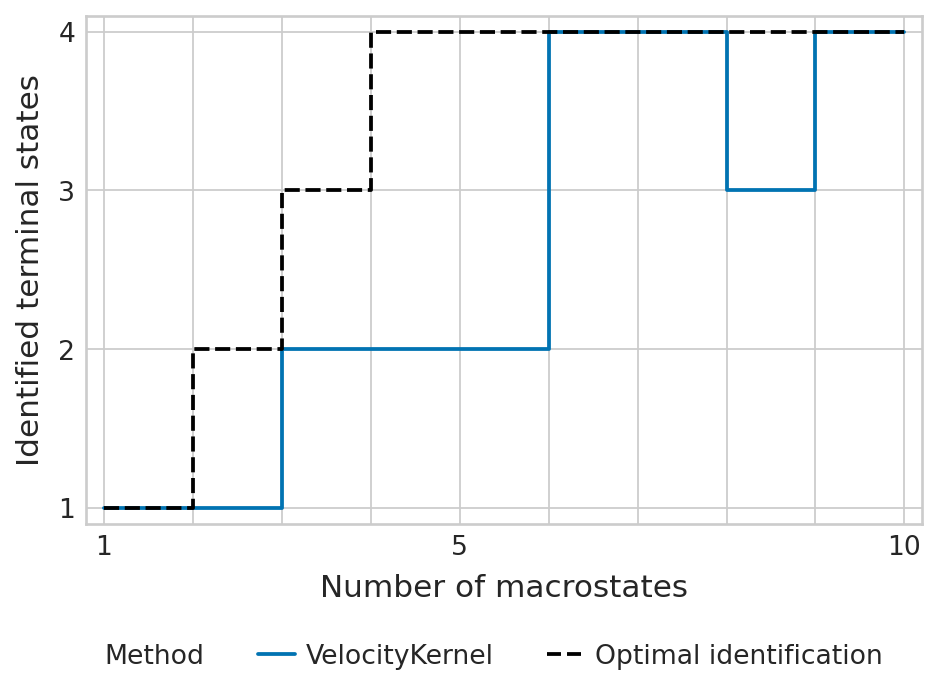
tsi_score = estimator.tsi(n_macrostates=10, terminal_states=terminal_states, cluster_key=cluster_key)
else:
tsi_score = estimator.tsi(n_macrostates=10, terminal_states=terminal_states, cluster_key=cluster_key)
estimator._tsi.to_df().to_csv(DATA_DIR / "mef" / "results" / "tsi-vk.csv", index=False)
print(f"TSI score: {tsi_score:.2f}")
TSI score: 0.79
/vol/storage/miniconda3/envs/cr2-py38/lib/python3.8/site-packages/anndata/_core/anndata.py:121: ImplicitModificationWarning: Transforming to str index.
warnings.warn("Transforming to str index.", ImplicitModificationWarning)
# For nice name in figure legend
estimator.kernel.__class__.__name__ = "VelocityKernel"
palette = {"VelocityKernel": "#0173b2", "Optimal identification": "#000000"}
if SAVE_FIGURES:
fpath = FIG_DIR / "realtime_kernel" / "mef" / f"tsi-vk.{FIGURE_FORMAT}"
else:
fpath = None
with mplscience.style_context():
sns.set_style(style="whitegrid")
estimator.plot_tsi(palette=palette, save=fpath)
plt.show()