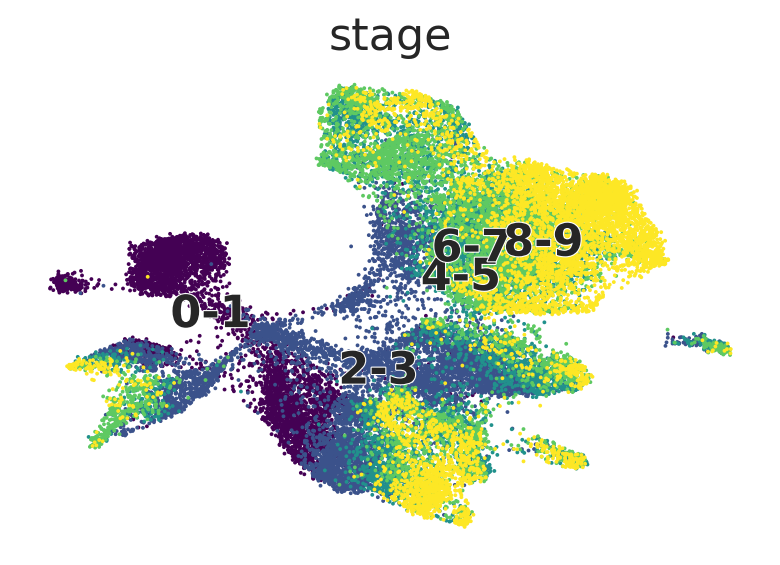
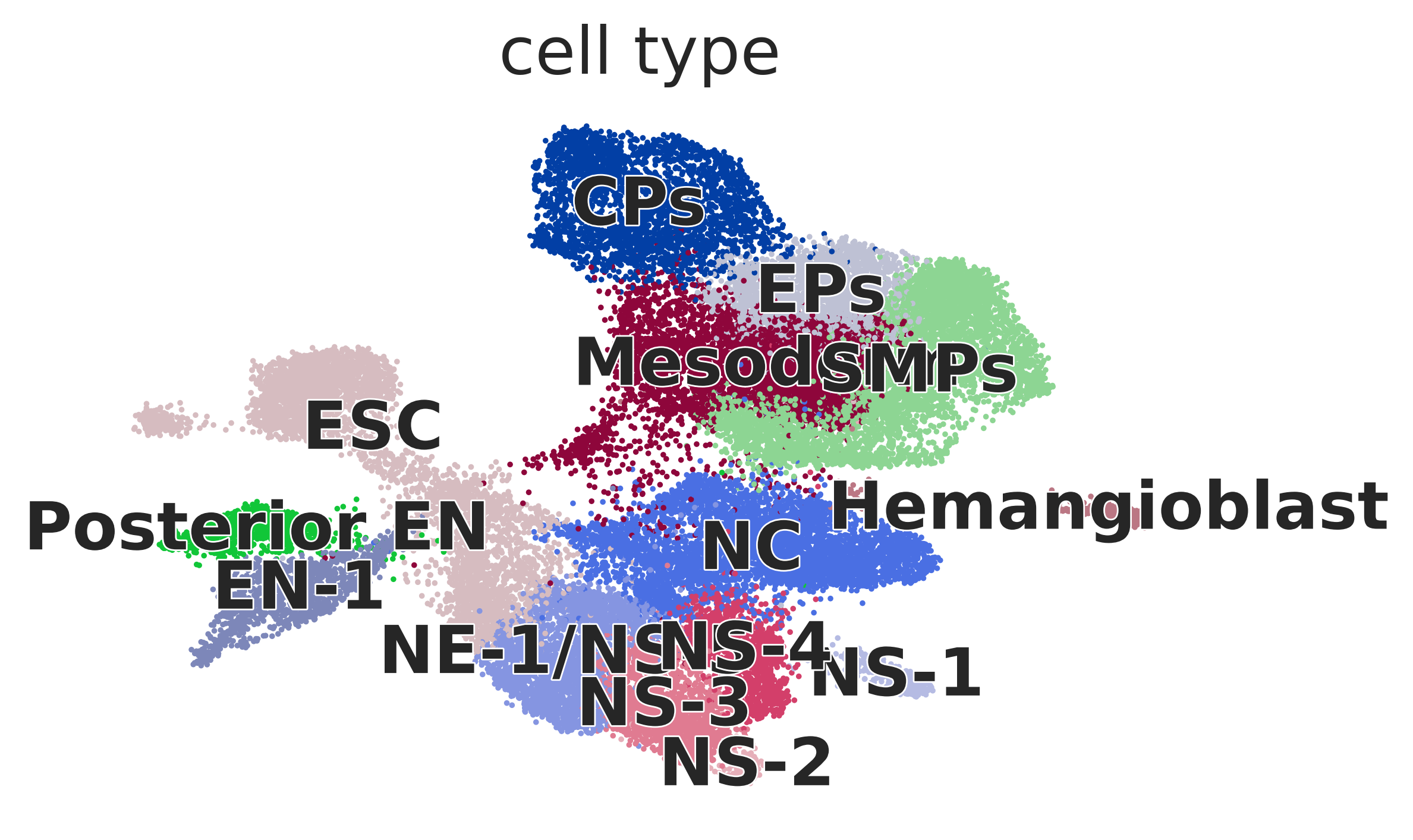
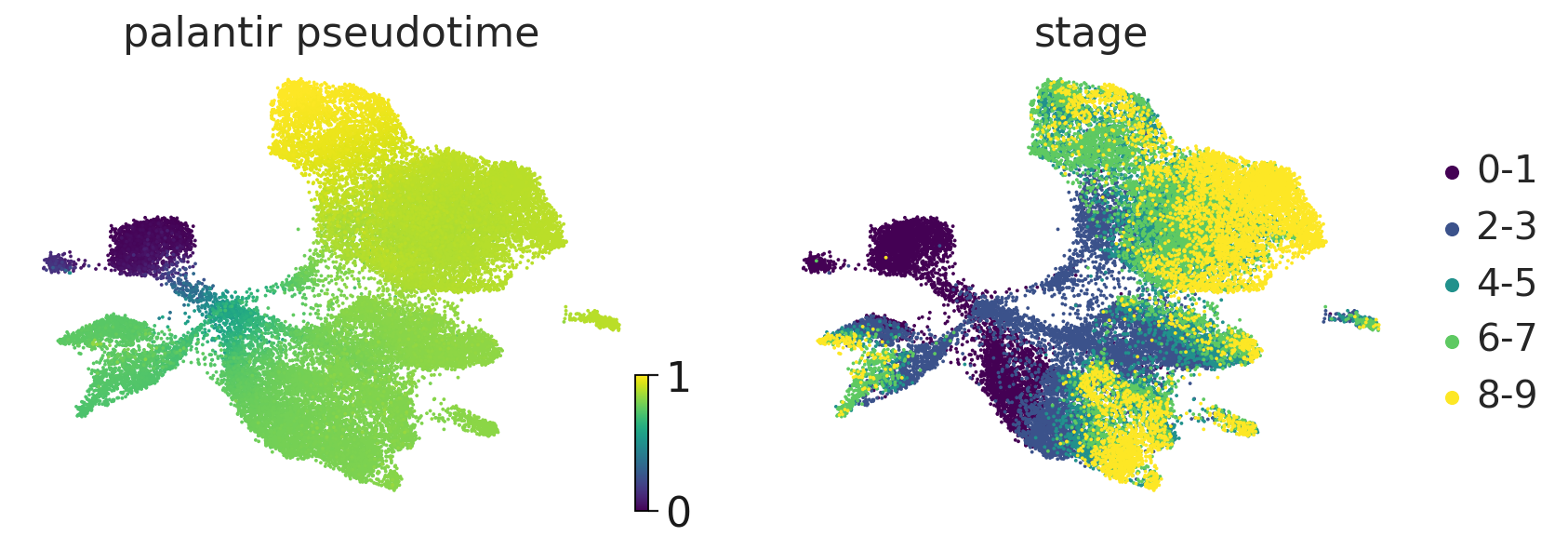
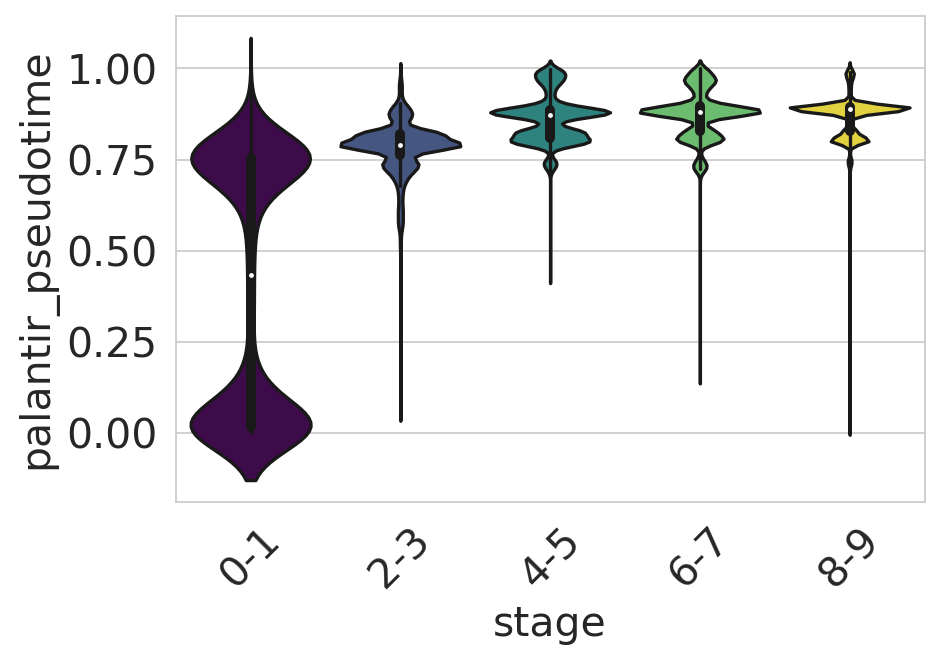
Palantir pseudotime#
Library imports#
import sys
import pandas as pd
import matplotlib as mpl
import matplotlib.pyplot as plt
import seaborn as sns
import palantir
import scanpy as sc
import scvelo as scv
sys.path.extend(["../../../", "."])
from paths import DATA_DIR, FIG_DIR # isort: skip # noqa: E402
findfont: Font family ['Raleway'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Lato'] not found. Falling back to DejaVu Sans.
Global seed set to 0
General settings#
sc.settings.verbosity = 2
scv.settings.verbosity = 3
mpl.use("module://matplotlib_inline.backend_inline")
mpl.rcParams["backend"]
plt.rcParams["font.family"] = "sans-serif"
plt.rcParams["font.sans-serif"] = ["Arial"]
scv.settings.set_figure_params("scvelo", dpi_save=400, dpi=80, transparent=True, fontsize=20, color_map="viridis")
SAVE_FIGURES = False
if SAVE_FIGURES:
(FIG_DIR / "cytotrace_kernel" / "embryoid_body").mkdir(parents=True, exist_ok=True)
FIGURE_FORMAT = "pdf"
Data loading#
adata = sc.read(DATA_DIR / "embryoid_body" / "embryoid_body.h5ad")
adata
AnnData object with n_obs × n_vars = 31029 × 19122
obs: 'stage', 'n_genes_by_counts', 'total_counts', 'total_counts_mt', 'pct_counts_mt', 'leiden', 'cell_type'
var: 'n_cells', 'mt', 'n_cells_by_counts', 'mean_counts', 'pct_dropout_by_counts', 'total_counts', 'highly_variable', 'means', 'dispersions', 'dispersions_norm'
uns: 'hvg', 'leiden', 'log1p', 'neighbors', 'pca', 'umap'
obsm: 'X_pca', 'X_umap'
varm: 'PCs'
obsp: 'connectivities', 'distances'
Pseudotime construction#
pc_projection = pd.DataFrame(adata.obsm["X_pca"].copy(), index=adata.obs_names)
diff_maps = palantir.utils.run_diffusion_maps(pc_projection, n_components=5)
Determing nearest neighbor graph...
computing neighbors
finished (0:02:35)
multiscale_space = palantir.utils.determine_multiscale_space(diff_maps)
magic_imputed = palantir.utils.run_magic_imputation(adata, diff_maps)
# See DPT notebook for root cell identification
root_idx = 1458
palantir_res = palantir.core.run_palantir(
multiscale_space, adata.obs_names[root_idx], use_early_cell_as_start=True, num_waypoints=500
)
Sampling and flocking waypoints...
Time for determining waypoints: 0.08258782227834066 minutes
Determining pseudotime...
Shortest path distances using 30-nearest neighbor graph...
findfont: Font family ['Raleway'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Lato'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Raleway'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Raleway'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Raleway'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Raleway'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Lato'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Lato'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Raleway'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Lato'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Lato'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Lato'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Raleway'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Raleway'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Lato'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Raleway'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Raleway'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Lato'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Raleway'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Lato'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Lato'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Lato'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Raleway'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Raleway'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Lato'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Lato'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Raleway'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Lato'] not found. Falling back to DejaVu Sans.
Time for shortest paths: 0.5515306274096171 minutes
Iteratively refining the pseudotime...
Correlation at iteration 1: 1.0000
Entropy and branch probabilities...
Markov chain construction...
Identification of terminal states...
Computing fundamental matrix and absorption probabilities...
Project results to all cells...
adata.obs["palantir_pseudotime"] = palantir_res.pseudotime
scv.pl.scatter(
adata,
c=["palantir_pseudotime", "stage"],
basis="umap",
legend_loc="right",
color_map="viridis",
)
if SAVE_FIGURES:
fig, ax = plt.subplots(figsize=(6, 4))
scv.pl.scatter(
adata, basis="umap", c="palantir_pseudotime", title="", legend_loc=False, colorbar=False, cmap="gnuplot2", ax=ax
)
fig.savefig(
FIG_DIR / "cytotrace_kernel" / "embryoid_body" / f"umap_colored_by_palantir_pseudotime.{FIGURE_FORMAT}",
format=FIGURE_FORMAT,
transparent=True,
bbox_inches="tight",
)
df = adata.obs[["palantir_pseudotime", "stage"]].copy()
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(6, 4))
sns.violinplot(
data=df,
x="stage",
y="palantir_pseudotime",
scale="width",
palette=["#440154", "#3b528b", "#21918c", "#5ec962", "#fde725"],
ax=ax,
)
ax.tick_params(axis="x", rotation=45)
ax.set_yticks([0, 0.25, 0.5, 0.75, 1])
plt.show()
if SAVE_FIGURES:
ax.set(xlabel=None, xticklabels=[], ylabel=None, yticklabels=[])
fig.savefig(
FIG_DIR / "cytotrace_kernel" / "embryoid_body" / f"palantir_vs_stage.{FIGURE_FORMAT}",
format=FIGURE_FORMAT,
transparent=True,
bbox_inches="tight",
)
sns.reset_orig()