Intestinal organoid differentiation - Velocity pseudotime#
Construct pseudotime based on transition matrix inferred from RNA velocity vector field on scEU-seq organoid data.
Library imports#
import os
import sys
import pandas as pd
from scipy.sparse import csr_matrix
import matplotlib.pyplot as plt
import seaborn as sns
import cellrank as cr
import scanpy as sc
import scvelo as scv
from scanpy.tools._dpt import DPT
from cr2 import get_symmetric_transition_matrix, running_in_notebook
sys.path.extend(["../../../", "."])
from paths import DATA_DIR, FIG_DIR # isort: skip # noqa: E402
Global seed set to 0
General settings#
sc.settings.verbosity = 3
scv.settings.verbosity = 3
cr.settings.verbosity = 2
scv.settings.set_figure_params("scvelo")
SAVE_FIGURES = False
if SAVE_FIGURES:
os.makedirs(FIG_DIR / "labeling_kernel", exist_ok=True)
Data loading#
adata = sc.read(DATA_DIR / "sceu_organoid" / "processed" / "preprocessed.h5ad")
adata
AnnData object with n_obs × n_vars = 3452 × 2000
obs: 'experiment', 'labeling_time', 'cell_type', 'som_cluster_id', 'cell_type_merged', 'initial_size', 'n_counts'
var: 'ensum_id', 'gene_count_corr', 'means', 'dispersions', 'dispersions_norm', 'highly_variable'
uns: 'cell_type_colors', 'neighbors', 'pca', 'umap'
obsm: 'X_pca', 'X_umap', 'X_umap_paper'
varm: 'PCs'
layers: 'labeled', 'total', 'unlabeled'
obsp: 'connectivities', 'distances'
Data preprocessing#
adata.obs["cell_type_merged"] = adata.obs["cell_type"].copy()
adata.obs["cell_type_merged"].replace({"Enteroendocrine cells": "Enteroendocrine progenitors"}, inplace=True)
celltype_to_color = dict(zip(adata.obs["cell_type"].cat.categories, adata.uns["cell_type_colors"]))
adata.uns["cell_type_merged_colors"] = list(
{cell_type: celltype_to_color[cell_type] for cell_type in adata.obs["cell_type_merged"].cat.categories}.values()
)
adata.layers["labeled_smoothed"] = csr_matrix.dot(adata.obsp["connectivities"], adata.layers["labeled"]).A
Velocity estimation#
alpha = pd.read_csv(DATA_DIR / "sceu_organoid" / "results" / "alpha.csv", index_col=0)
alpha.index = alpha.index.astype(str)
adata.layers["transcription_rate"] = alpha.loc[adata.obs_names, adata.var_names]
gamma = pd.read_csv(DATA_DIR / "sceu_organoid" / "results" / "gamma.csv", index_col=0)
gamma.index = gamma.index.astype(str)
adata.layers["degradation_rate"] = alpha.loc[adata.obs_names, adata.var_names]
adata.layers["velocity_labeled"] = (alpha - gamma * adata.layers["labeled_smoothed"]).values
Pseudotime construction#
vk = cr.kernels.VelocityKernel(adata, xkey="labeled_smoothed", vkey="velocity_labeled").compute_transition_matrix()
ck = cr.kernels.ConnectivityKernel(adata).compute_transition_matrix()
combined_kernel = 0.8 * vk + 0.2 * ck
Computing transition matrix using `'deterministic'` model
Using `softmax_scale=2.4573`
Finish (0:00:04)
Computing transition matrix based on `adata.obsp['connectivities']`
Finish (0:00:00)
dpt = DPT(adata=adata, neighbors_key="neighbors")
dpt._transitions_sym = get_symmetric_transition_matrix(combined_kernel.transition_matrix)
dpt.compute_eigen(n_comps=15, random_state=0)
adata.obsm["X_diffmap"] = dpt.eigen_basis
adata.uns["diffmap_evals"] = dpt.eigen_values
eigenvalues of transition matrix
[1.0024191 0.98475575 0.9499719 0.9143363 0.90075344 0.87188613
0.8576526 0.81567115 0.7717359 0.76253957 0.74204767 0.7327005
0.72588813 0.7087067 0.6973775 ]
adata.uns["iroot"] = 2899 # See dpt.ipynb
sc.tl.dpt(adata)
computing Diffusion Pseudotime using n_dcs=10
finished: added
'dpt_pseudotime', the pseudotime (adata.obs) (0:00:00)
adata.obs.rename({"dpt_pseudotime": "velocity_pseudotime"}, axis=1, inplace=True)
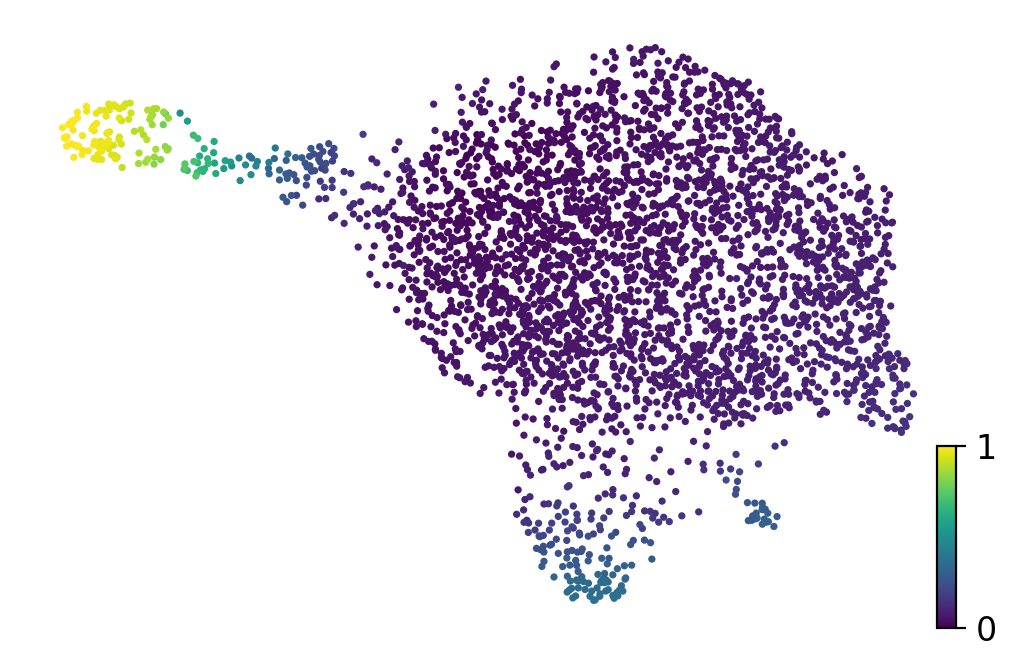
if running_in_notebook():
fig, ax = plt.subplots(figsize=(6, 4))
scv.pl.scatter(adata, basis="umap", color="velocity_pseudotime", title="", color_map="viridis", ax=ax)
if SAVE_FIGURES:
fig, ax = plt.subplots(figsize=(6, 4))
scv.pl.scatter(
adata, basis="umap", color="velocity_pseudotime", title="", color_map="viridis", colorbar=False, ax=ax
)
fig.savefig(
FIG_DIR / "labeling_kernel" / "umap_colored_by_velocity_pseudotime.eps",
format="eps",
transparent=True,
bbox_inches="tight",
)
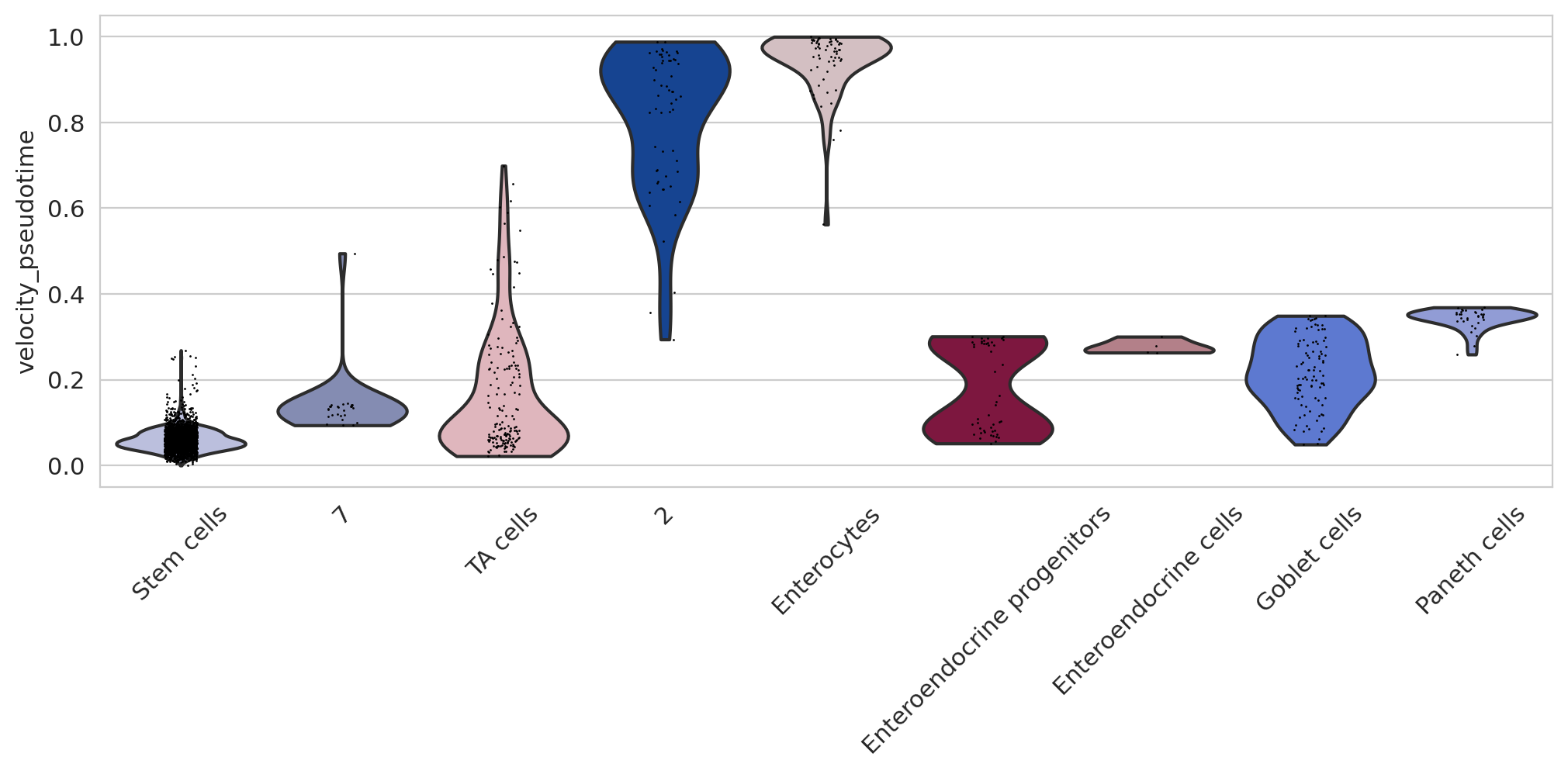
if running_in_notebook():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(12, 4))
sc.pl.violin(
adata,
keys=["velocity_pseudotime"],
groupby="cell_type",
rotation=45,
title="",
legend_loc="none",
order=[
"Stem cells",
"7",
"TA cells",
"2",
"Enterocytes",
"Enteroendocrine progenitors",
"Enteroendocrine cells",
"Goblet cells",
"Paneth cells",
],
ax=ax,
)
if SAVE_FIGURES:
ax.set(xticklabels=[])
fig.savefig(
FIG_DIR / "labeling_kernel" / "velocity_pseudotime_vs_cell_type.eps",
format="eps",
transparent=True,
bbox_inches="tight",
)
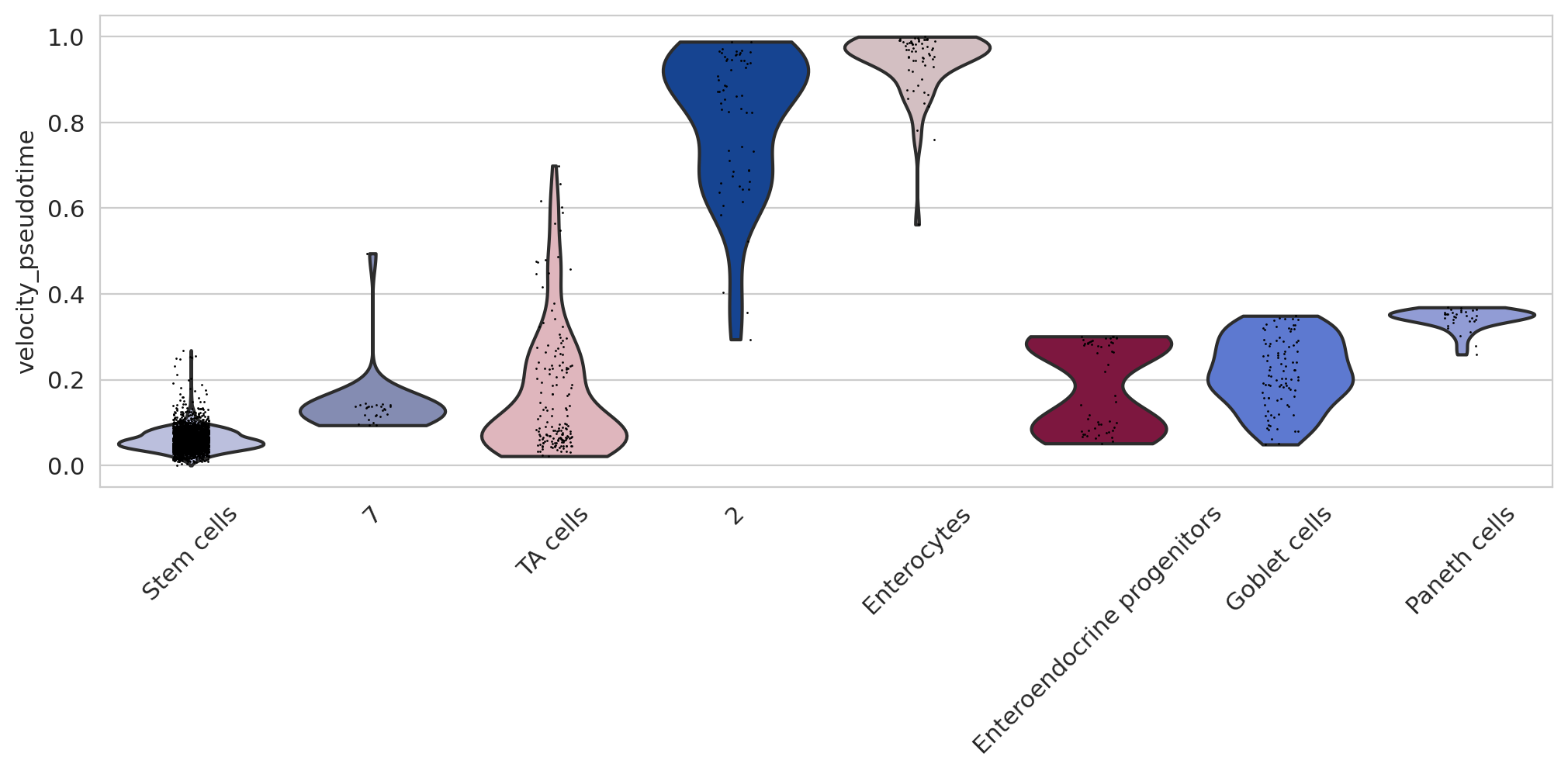
if running_in_notebook():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(12, 4))
sc.pl.violin(
adata,
keys=["velocity_pseudotime"],
groupby="cell_type_merged",
rotation=45,
title="",
legend_loc="none",
order=[
"Stem cells",
"7",
"TA cells",
"2",
"Enterocytes",
"Enteroendocrine progenitors",
"Goblet cells",
"Paneth cells",
],
ax=ax,
)
if SAVE_FIGURES:
ax.set(xticklabels=[])
fig.savefig(
FIG_DIR / "labeling_kernel" / "velocity_pseudotime_vs_cell_type_merged.eps",
format="eps",
transparent=True,
bbox_inches="tight",
)
adata.obs["velocity_pseudotime"].to_csv(DATA_DIR / "sceu_organoid" / "results" / "velocity_pseudotime.csv")