VelocityKernel vs. RealTimeKernel - TSI#
Library imports#
%load_ext autoreload
%autoreload 2
import sys
import pandas as pd
import matplotlib.pyplot as plt
import mplscience
import seaborn as sns
from cr2.analysis import plot_tsi
sys.path.extend(["../../../", "."])
from paths import DATA_DIR, FIG_DIR # isort: skip # noqa: E402
Global seed set to 0
General settings#
SAVE_FIGURES = False
if SAVE_FIGURES:
(FIG_DIR / "realtime_kernel" / "pharyngeal_endoderm").mkdir(parents=True, exist_ok=True)
FIGURE_FORMAT = "pdf"
Constants#
Data loading#
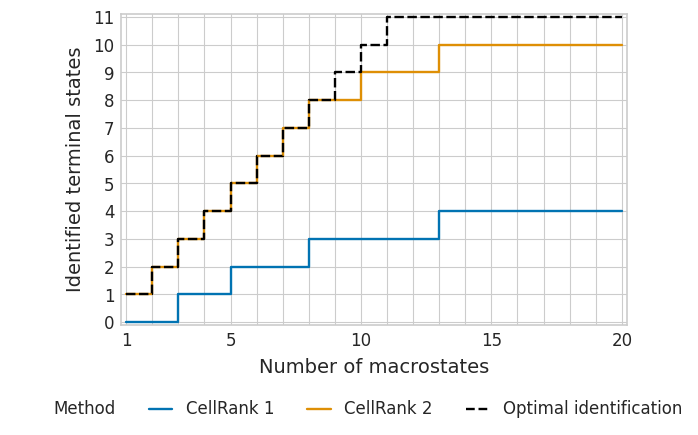
tsi_cr1_full = pd.read_csv(DATA_DIR / "pharyngeal_endoderm" / "results" / "tsi-full_data-vk.csv")
tsi_cr1_full.head()
| number_of_macrostates | identified_terminal_states | optimal_identification | |
|---|---|---|---|
| 0 | 20 | 4 | 11 |
| 1 | 19 | 4 | 11 |
| 2 | 18 | 4 | 11 |
| 3 | 17 | 4 | 11 |
| 4 | 16 | 4 | 11 |
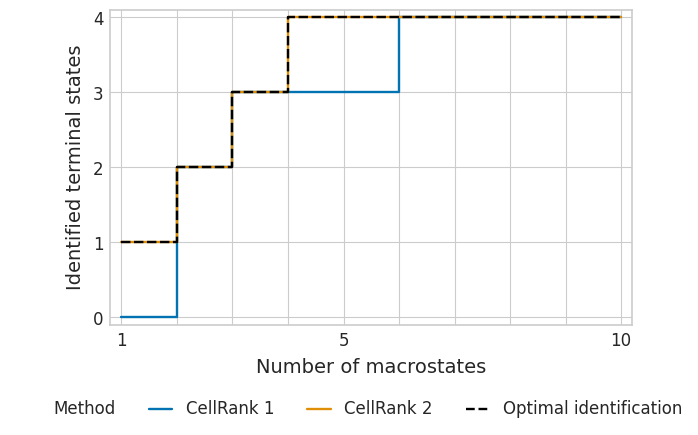
tsi_cr1_subset = pd.read_csv(DATA_DIR / "pharyngeal_endoderm" / "results" / "tsi-subsetted_data-vk.csv")
tsi_cr1_subset.head()
| number_of_macrostates | identified_terminal_states | optimal_identification | |
|---|---|---|---|
| 0 | 10 | 4 | 4 |
| 1 | 9 | 4 | 4 |
| 2 | 8 | 4 | 4 |
| 3 | 7 | 4 | 4 |
| 4 | 6 | 4 | 4 |
tsi_cr2_full = pd.read_csv(DATA_DIR / "pharyngeal_endoderm" / "results" / "tsi-full_data-rtk.csv")
tsi_cr2_full.head()
| number_of_macrostates | identified_terminal_states | optimal_identification | |
|---|---|---|---|
| 0 | 1 | 1 | 1 |
| 1 | 2 | 2 | 2 |
| 2 | 3 | 3 | 3 |
| 3 | 4 | 4 | 4 |
| 4 | 5 | 5 | 5 |
tsi_cr2_subset = pd.read_csv(DATA_DIR / "pharyngeal_endoderm" / "results" / "tsi-subsetted_data-rtk.csv")
tsi_cr2_subset.head()
| number_of_macrostates | identified_terminal_states | optimal_identification | |
|---|---|---|---|
| 0 | 10 | 4 | 4 |
| 1 | 9 | 4 | 4 |
| 2 | 8 | 4 | 4 |
| 3 | 7 | 4 | 4 |
| 4 | 6 | 4 | 4 |
Data preprocessing#
tsi_cr1_full["method"] = "CellRank 1"
tsi_cr1_subset["method"] = "CellRank 1"
tsi_cr2_full["method"] = "CellRank 2"
tsi_cr2_subset["method"] = "CellRank 2"
df_full = pd.concat([tsi_cr1_full, tsi_cr2_full])
df_subset = pd.concat([tsi_cr1_subset, tsi_cr2_subset])
Plotting#
palette = {"CellRank 1": "#0173b2", "CellRank 2": "#DE8F05", "Optimal identification": "#000000"}
if SAVE_FIGURES:
fname = FIG_DIR / "realtime_kernel" / "pharyngeal_endoderm" / f"tsi_ranking-full_data.{FIGURE_FORMAT}"
else:
fname = None
with mplscience.style_context():
sns.set_style(style="whitegrid")
plot_tsi(df=df_full, palette=palette, fname=fname)
plt.show()
palette = {"CellRank 1": "#0173b2", "CellRank 2": "#DE8F05", "Optimal identification": "#000000"}
if SAVE_FIGURES:
fname = FIG_DIR / "realtime_kernel" / "pharyngeal_endoderm" / f"tsi_ranking-subsetted_data.{FIGURE_FORMAT}"
else:
fname = None
with mplscience.style_context():
sns.set_style(style="whitegrid")
plot_tsi(df=df_subset, palette=palette, fname=fname)
plt.show()