Performance comparison of inference on cell cycle#
Notebook compares metrics for velocity, latent time and GRN inference across different methods applied to cell cycle data.
Library imports#
import pandas as pd
import matplotlib.pyplot as plt
import mplscience
import seaborn as sns
from rgv_tools import DATA_DIR, FIG_DIR
General settings#
DATASET = "cell_cycle"
SAVE_FIGURES = False
if SAVE_FIGURES:
(FIG_DIR / DATASET).mkdir(parents=True, exist_ok=True)
FIGURE_FORMATE = "svg"
Constants#
TIME_METHODS = [
"regvelo",
"velovi",
"scvelo",
"unitvelo",
"velovae_vae",
"velovae_fullvb",
"cell2fate",
"tfvelo",
"dpt",
]
VELO_METHODS = ["regvelo", "velovi", "scvelo", "unitvelo", "velovae_vae", "velovae_fullvb", "cell2fate", "tfvelo"]
GRN_METHODS = ["regvelo_grn", "grnboost2", "celloracle", "correlation", "splicejac", "tfvelo_grn"]
Data loading#
time_df = []
cbc_df = []
confi_df = []
grn_df = []
for method in TIME_METHODS:
df = pd.read_parquet(DATA_DIR / DATASET / "results" / f"{method}_correlation.parquet")
df.columns = f"{method}_" + df.columns
df = df.mean(0)
time_df.append(df)
for method in VELO_METHODS:
df = pd.read_parquet(DATA_DIR / DATASET / "results" / f"{method}_cbc.parquet")
del df["State transition"]
df.columns = f"{method}_" + df.columns
df = df.mean(0)
cbc_df.append(df)
for method in VELO_METHODS:
df = pd.read_parquet(DATA_DIR / DATASET / "results" / f"{method}_confidence.parquet")
df.columns = f"{method}_" + df.columns
confi_df.append(df)
for method in GRN_METHODS:
df = pd.read_parquet(DATA_DIR / DATASET / "results" / f"{method}_correlation.parquet")
df = pd.DataFrame({"AUROC": df.iloc[0, 0]})
df.columns = f"{method}_" + df.columns
grn_df.append(df)
time_df = pd.concat(time_df).reset_index()
time_df.columns = ["method", "spearman_corr"]
cbc_df = pd.concat(cbc_df).reset_index()
cbc_df.columns = ["method", "CBC"]
confi_df = pd.concat(confi_df, axis=1).melt(var_name="method", value_name="confidence")
grn_df = pd.concat(grn_df, axis=1).melt(var_name="method", value_name="AUROC")
Processing results table#
time_df.iloc[:, 0] = time_df.iloc[:, 0].str.removesuffix("_time")
cbc_df.iloc[:, 0] = cbc_df.iloc[:, 0].str.removesuffix("_CBC")
confi_df.iloc[:, 0] = confi_df.iloc[:, 0].str.removesuffix("_velocity_confidence")
grn_df.iloc[:, 0] = grn_df.iloc[:, 0].str.removesuffix("_AUROC")
Analysis#
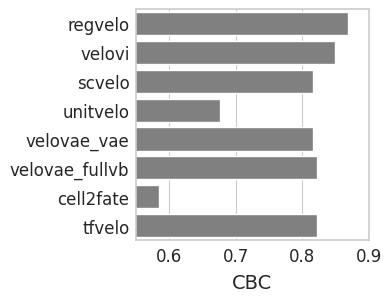
CBC#
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(3, 3), sharey=True)
# Plot the second Seaborn plot on the first subplot
sns.barplot(y="method", x="CBC", data=cbc_df, capsize=0.1, color="grey", order=VELO_METHODS, ax=ax)
ax.set_xlabel("CBC", fontsize=14)
ax.set_ylabel("")
plt.xlim(0.55, 0.9)
plt.show()
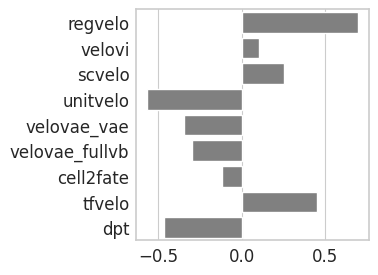
Latent time#
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(3, 3), sharey=True)
# Plot the second Seaborn plot on the first subplot
sns.barplot(y="method", x="spearman_corr", data=time_df, capsize=0.1, color="grey", order=TIME_METHODS, ax=ax)
ax.set_xlabel("", fontsize=14)
ax.set_ylabel("")
plt.show()
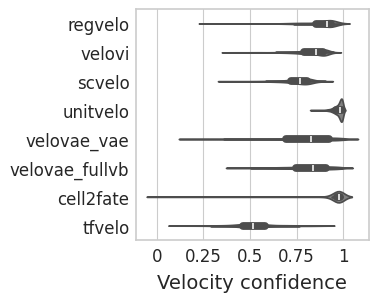
Confidence#
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(3, 3), sharey=True)
sns.violinplot(
data=confi_df,
ax=ax,
orient="h",
y="method",
x="confidence",
color="grey",
order=VELO_METHODS,
)
# plt.legend(title='', loc='lower center', bbox_to_anchor=(0.5, -0.6), ncol=3)
ax.set_xticks([0, 0.25, 0.5, 0.75, 1])
ax.set_xticklabels([0, 0.25, 0.5, 0.75, 1])
ax.set_xlabel("Velocity confidence", fontsize=14)
ax.set_ylabel("")
plt.show()
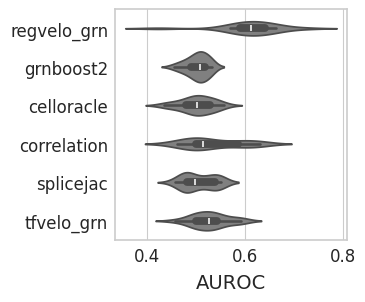
GRN#
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(3, 3), sharey=True)
sns.violinplot(
data=grn_df,
ax=ax,
orient="h",
y="method",
x="AUROC",
color="grey",
order=GRN_METHODS,
)
# plt.legend(title='', loc='lower center', bbox_to_anchor=(0.5, -0.6), ncol=3)
ax.set_xlabel("AUROC", fontsize=14)
ax.set_ylabel("")
plt.show()