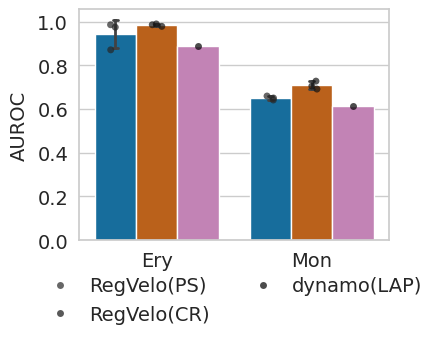
Comparison of driver TF ranking performance#
Notebook compares the performance of RegVelo (CR), RegVelo (perturbation), and Dynamo in predicting driver genes.
Library imports#
import numpy as np
import pandas as pd
from sklearn.metrics import roc_auc_score
import matplotlib.pyplot as plt
import mplscience
import seaborn as sns
import cellrank as cr
import scanpy as sc
from regvelo import REGVELOVI
from rgv_tools import DATA_DIR, FIG_DIR
from rgv_tools.benchmarking import set_output
from rgv_tools.core import METHOD_PALETTE_DRIVER
from rgv_tools.perturbation import aggregate_model_predictions
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_csv from `anndata` is deprecated. Import anndata.io.read_csv instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_excel from `anndata` is deprecated. Import anndata.io.read_excel instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_hdf from `anndata` is deprecated. Import anndata.io.read_hdf instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_loom from `anndata` is deprecated. Import anndata.io.read_loom instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_mtx from `anndata` is deprecated. Import anndata.io.read_mtx instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_text from `anndata` is deprecated. Import anndata.io.read_text instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_umi_tools from `anndata` is deprecated. Import anndata.io.read_umi_tools instead.
warnings.warn(msg, FutureWarning)
Constants#
DATASET = "hematopoiesis"
SAVE_DATA = True
if SAVE_DATA:
(DATA_DIR / DATASET / "results").mkdir(parents=True, exist_ok=True)
SAVE_FIGURES = True
if SAVE_FIGURES:
(FIG_DIR / DATASET).mkdir(parents=True, exist_ok=True)
Mon_driver = ["SPI1", "TCF4", "STAT6", "MEF2C"]
Ery_driver = ["NFIA", "GATA1", "TAL1", "GFI1B", "LMO2"]
terminal_states = ["Meg", "Mon", "Bas", "Ery"]
Data loading#
adata = sc.read_h5ad(DATA_DIR / DATASET / "processed" / "adata_preprocessed.h5ad")
TF = adata.var_names[adata.var["TF"]]
regvelo_prediction = aggregate_model_predictions(DATA_DIR / DATASET / "results")
HSC_Mon_ranking = pd.read_csv(DATA_DIR / DATASET / "results" / "HSC_Mon_ranking.csv", index_col=0)
HSC_Ery_ranking = pd.read_csv(DATA_DIR / DATASET / "results" / "HSC_Ery_ranking.csv", index_col=0)
RegVelo’s in silico perturbation driver identification#
ery_auc_rgv = []
mon_auc_rgv = []
for coef in regvelo_prediction:
## ranking Erythroid drivers
driver = Ery_driver
cell_fate = "Ery"
ID = set(driver).intersection(set(coef.index.tolist()))
label = np.zeros(coef.shape[0])
label[[i for i in range(coef.shape[0]) if coef.index.tolist()[i] in list(ID)]] = 1
score = coef.copy().loc[:, cell_fate]
score[np.isnan(score)] = 0
ery_auc_rgv.append(roc_auc_score(label, (score)))
## ranking Monocyte drivers
driver = Mon_driver
cell_fate = "Mon"
ID = set(driver).intersection(set(coef.index.tolist()))
label = np.zeros(coef.shape[0])
label[[i for i in range(coef.shape[0]) if coef.index.tolist()[i] in list(ID)]] = 1
score = coef.copy().loc[:, cell_fate]
score[np.isnan(score)] = 0
mon_auc_rgv.append(roc_auc_score(label, (score)))
CellRank’s driver identification#
ery_auc_cr = []
mon_auc_cr = []
## Ery driver ranking
for method in ["0", "1", "2"]:
vae = REGVELOVI.load(DATA_DIR / DATASET / "processed" / "perturb_repeat_runs" / f"rgv_model_{method}", adata)
set_output(adata, vae, n_samples=30, batch_size=adata.n_obs)
## Using CellRank identify driver
vk = cr.kernels.VelocityKernel(adata)
vk.compute_transition_matrix()
ck = cr.kernels.ConnectivityKernel(adata).compute_transition_matrix()
g = cr.estimators.GPCCA(0.8 * vk + 0.2 * ck)
## evaluate the fate prob on original space
g.compute_macrostates(n_states=6, cluster_key="cell_type")
g.set_terminal_states(terminal_states)
g.compute_fate_probabilities()
df = g.compute_lineage_drivers(cluster_key="cell_type")
df = df.loc[TF, :].copy()
driver = Ery_driver
cell_fate = "Ery" + "_corr"
ID = set(driver).intersection(set(df.index.tolist()))
label = np.zeros(df.shape[0])
label[[i for i in range(df.shape[0]) if df.index.tolist()[i] in list(ID)]] = 1
# Calculate AUROC
score_raw = df.copy().loc[:, cell_fate]
score_raw[np.isnan(score_raw)] = 0
auroc_ery = roc_auc_score(label, (score_raw))
ery_auc_cr.append(auroc_ery)
## Mon driver ranking
driver = Mon_driver
cell_fate = "Mon" + "_corr"
ID = set(driver).intersection(set(df.index.tolist()))
label = np.zeros(df.shape[0])
label[[i for i in range(df.shape[0]) if df.index.tolist()[i] in list(ID)]] = 1
# Calculate AUROC
score_raw = df.copy().loc[:, cell_fate]
score_raw[np.isnan(score_raw)] = 0
auroc_mon = roc_auc_score(label, (score_raw))
mon_auc_cr.append(auroc_mon)
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/hematopoiesis/processed/perturb_repeat_runs/rgv_model_0
/model.pt already downloaded
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:168: PossibleUserWarning: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
rank_zero_warn(
100%|██████████| 1947/1947 [00:24<00:00, 80.61cell/s]
100%|██████████| 1947/1947 [00:00<00:00, 2979.26cell/s]
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
WARNING: Unable to import petsc4py. For installation, please refer to: https://petsc4py.readthedocs.io/en/stable/install.html.
Defaulting to `'gmres'` solver.
100%|██████████| 4/4 [00:00<00:00, 74.20/s]
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/hematopoiesis/processed/perturb_repeat_runs/rgv_model_1
/model.pt already downloaded
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:168: PossibleUserWarning: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
rank_zero_warn(
100%|██████████| 1947/1947 [00:00<00:00, 3034.55cell/s]
100%|██████████| 1947/1947 [00:00<00:00, 3069.80cell/s]
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
WARNING: Using `6` components would split a block of complex conjugate eigenvalues. Using `n_components=7`
WARNING: Unable to compute macrostates with `n_states=6` because it will split complex conjugate eigenvalues. Using `n_states=7`
100%|██████████| 4/4 [00:00<00:00, 67.83/s]
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/hematopoiesis/processed/perturb_repeat_runs/rgv_model_2
/model.pt already downloaded
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:168: PossibleUserWarning: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
rank_zero_warn(
100%|██████████| 1947/1947 [00:00<00:00, 2493.91cell/s]
100%|██████████| 1947/1947 [00:00<00:00, 2532.22cell/s]
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
100%|██████████| 4/4 [00:00<00:00, 61.69/s]
Dynamo’s driver identification#
HSC_Ery_ranking["filter"] = [i in TF for i in list(HSC_Ery_ranking["all"])]
HSC_Mon_ranking["filter"] = [i in TF for i in list(HSC_Mon_ranking["all"])]
HSC_Ery_ranking = HSC_Ery_ranking.query("filter == True")
HSC_Mon_ranking = HSC_Mon_ranking.query("filter == True")
ID = set(Ery_driver).intersection(set(HSC_Ery_ranking.iloc[:, 0].tolist()))
label = np.zeros(HSC_Ery_ranking.shape[0])
label[[i for i in range(HSC_Ery_ranking.shape[0]) if HSC_Ery_ranking.iloc[:, 0].tolist()[i] in list(ID)]] = 1
score = -1 * np.array(range(HSC_Ery_ranking.shape[0]))
auroc_ery_dynamo = roc_auc_score(label, score)
print("AUROC:", auroc_ery_dynamo)
AUROC: 0.8888888888888888
ID = set(Mon_driver).intersection(set(HSC_Mon_ranking.iloc[:, 0].tolist()))
label = np.zeros(HSC_Mon_ranking.shape[0])
label[[i for i in range(HSC_Mon_ranking.shape[0]) if HSC_Mon_ranking.iloc[:, 0].tolist()[i] in list(ID)]] = 1
score = -1 * np.array(range(HSC_Mon_ranking.shape[0]))
auroc_mon_dynamo = roc_auc_score(label, score)
print("AUROC:", auroc_mon_dynamo)
AUROC: 0.6136363636363636
Plot driver ranking results#
data = pd.DataFrame(
{
"AUROC": ery_auc_rgv + mon_auc_rgv + ery_auc_cr + mon_auc_cr + [auroc_ery_dynamo, auroc_mon_dynamo],
"Terminal state": ["Ery"] * 3 + ["Mon"] * 3 + ["Ery"] * 3 + ["Mon"] * 3 + ["Ery", "Mon"],
"Method": ["RegVelo (PS)"] * 6 + ["RegVelo (CR)"] * 6 + ["dynamo (LAP)"] * 2,
}
)
with mplscience.style_context():
pal = METHOD_PALETTE_DRIVER
sns.set(style="whitegrid")
fig, ax = plt.subplots(figsize=(4, 3))
sns.barplot(x="Terminal state", y="AUROC", hue="Method", data=data, ci="sd", capsize=0.1, errwidth=2, palette=pal)
sns.stripplot(
x="Terminal state", y="AUROC", hue="Method", data=data, dodge=True, jitter=True, color="black", alpha=0.7
)
handles, labels = ax.get_legend_handles_labels()
ax.legend(handles[3:6], labels[3:6], bbox_to_anchor=(0.5, -0.44), loc="lower center", ncol=2, fontsize=14)
plt.ylabel("AUROC", fontsize=14)
plt.xlabel("", fontsize=14)
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
if SAVE_FIGURES:
plt.savefig(FIG_DIR / DATASET / "driver_ranking.svg", format="svg", transparent=True, bbox_inches="tight")
plt.show()