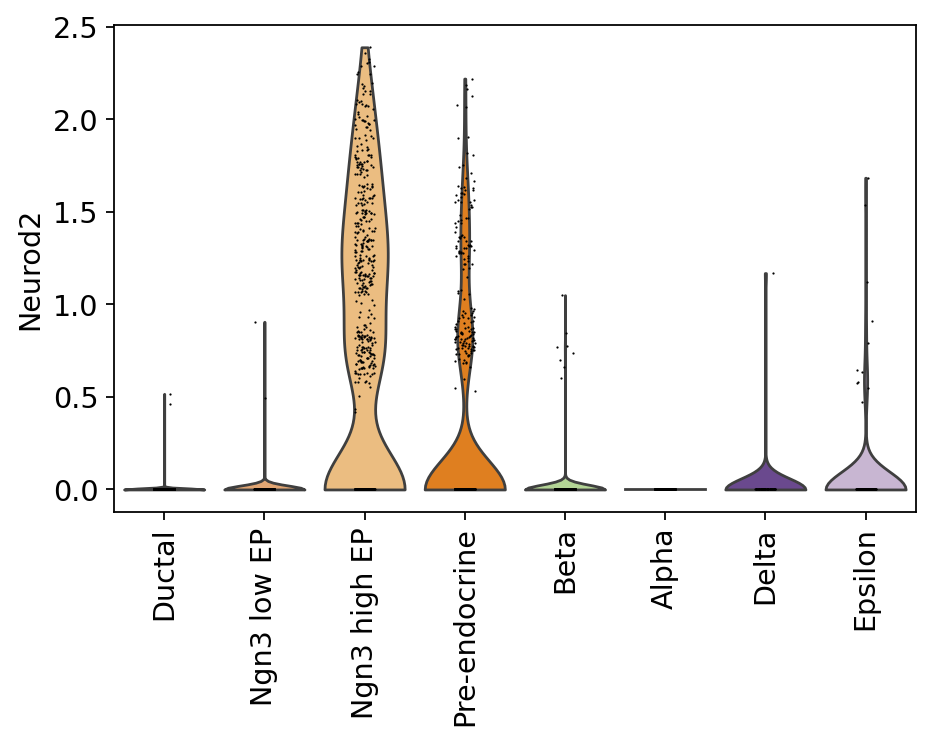
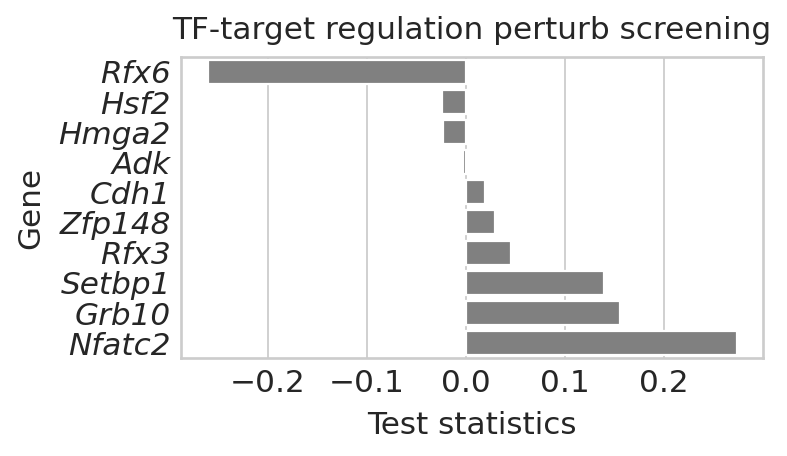
Neurod2 target analysis#
Notebooks analyses Neurod2 targets, identifying the Neurod2 - Rfx6 regulatory link for epsilon cells.
Library imports#
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import mplscience
import seaborn as sns
import scanpy as sc
import scvelo as scv
from regvelo import REGVELOVI
from rgv_tools import DATA_DIR, FIG_DIR
from rgv_tools.perturbation import (
aggregate_model_predictions,
get_list_name,
inferred_GRN,
RegulationScanning,
)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_csv from `anndata` is deprecated. Import anndata.io.read_csv instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_excel from `anndata` is deprecated. Import anndata.io.read_excel instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_hdf from `anndata` is deprecated. Import anndata.io.read_hdf instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_loom from `anndata` is deprecated. Import anndata.io.read_loom instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_mtx from `anndata` is deprecated. Import anndata.io.read_mtx instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_text from `anndata` is deprecated. Import anndata.io.read_text instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_umi_tools from `anndata` is deprecated. Import anndata.io.read_umi_tools instead.
warnings.warn(msg, FutureWarning)
General settings#
%matplotlib inline
plt.rcParams["svg.fonttype"] = "none"
sns.reset_defaults()
sns.reset_orig()
scv.settings.set_figure_params("scvelo", dpi_save=400, dpi=80, transparent=True, fontsize=14, color_map="viridis")
Constants#
DATASET = "pancreatic_endocrinogenesis"
SAVE_DATA = True
if SAVE_DATA:
(DATA_DIR / DATASET / "results").mkdir(parents=True, exist_ok=True)
(DATA_DIR / DATASET / "results" / "Neurod2_screening_repeat_runs").mkdir(parents=True, exist_ok=True)
SAVE_FIGURES = True
if SAVE_FIGURES:
(FIG_DIR / DATASET).mkdir(parents=True, exist_ok=True)
TERMINAL_STATES = ["Alpha", "Beta", "Delta", "Epsilon"]
Data loading#
adata = sc.read_h5ad(DATA_DIR / DATASET / "processed" / "adata_preprocessed.h5ad")
palette = dict(zip(adata.obs["clusters"].cat.categories, adata.uns["clusters_colors"]))
Build RegVelo’s GRN#
GRN_list = [] # Assuming there are 4 metrics
for i in range(5):
model_name = "rgv_model_" + str(i)
model = DATA_DIR / DATASET / "processed" / "perturb_repeat_runs" / model_name
### load model
vae = REGVELOVI.load(model, adata)
GRN_list.append(inferred_GRN(vae, adata, label="clusters", group="all"))
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_0/model.pt already downloaded
Computing global GRN...
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_1/model.pt already downloaded
Computing global GRN...
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_2/model.pt already downloaded
Computing global GRN...
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_3/model.pt already downloaded
Computing global GRN...
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_4/model.pt already downloaded
Computing global GRN...
grn_all = np.mean(np.stack(GRN_list), axis=0)
predict_weight = grn_all[:, [i == "Neurod2" for i in adata.var.index]]
prior = vae.module.v_encoder.mask_m_raw[:, [i == "Neurod2" for i in adata.var.index]].detach().cpu().numpy()
predict_weight = pd.DataFrame(predict_weight, index=adata.var.index)
predict_weight.loc[:, "weight"] = predict_weight.iloc[:, 0].abs()
predict_weight.loc[:, "prior"] = prior
Screening from top-10 targets#
genes = predict_weight.sort_values("weight", ascending=False).iloc[:10, :].index.tolist()
terminal_states = ["Alpha", "Delta", "Beta", "Epsilon"]
for i in range(5):
print("loading model...")
model_name = "rgv_model_" + str(i)
model = DATA_DIR / DATASET / "processed" / "perturb_repeat_runs" / model_name
coef_name = "coef_" + str(i)
res_save_coef = DATA_DIR / DATASET / "results" / "Neurod2_screening_repeat_runs" / coef_name
pval_name = "pval_" + str(i)
res_save_pval = DATA_DIR / DATASET / "results" / "Neurod2_screening_repeat_runs" / pval_name
print("inferring perturbation...")
perturb_screening = RegulationScanning(
model, adata, 8, "clusters", TERMINAL_STATES, "Neurod2", genes, 0, method="t-statistics"
)
coef = pd.DataFrame(np.array(perturb_screening["coefficient"]))
coef.index = perturb_screening["target"]
coef.columns = get_list_name(perturb_screening["coefficient"][0])
pval = pd.DataFrame(np.array(perturb_screening["pvalue"]))
pval.index = perturb_screening["target"]
pval.columns = get_list_name(perturb_screening["pvalue"][0])
coef.to_csv(res_save_coef)
pval.to_csv(res_save_pval)
loading model...
inferring perturbation...
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_0/model.pt already downloaded
2024-12-02 03:02:15,686 - INFO - Using pre-computed Schur decomposition
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_0/model.pt already downloaded
2024-12-02 03:03:20,561 - INFO - Using pre-computed Schur decomposition
Done Rfx3
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_0/model.pt already downloaded
2024-12-02 03:04:25,869 - INFO - Using pre-computed Schur decomposition
Done Adk
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_0/model.pt already downloaded
2024-12-02 03:05:35,107 - INFO - Using pre-computed Schur decomposition
Done Zfp148
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_0/model.pt already downloaded
2024-12-02 03:06:40,912 - INFO - Using pre-computed Schur decomposition
Done Nfatc2
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_0/model.pt already downloaded
[0]PETSC ERROR: ------------------------------------------------------------------------
[0]PETSC ERROR: Caught signal number 13 Broken Pipe: Likely while reading or writing to a socket
[0]PETSC ERROR: Try option -start_in_debugger or -on_error_attach_debugger
[0]PETSC ERROR: or see https://petsc.org/release/faq/#valgrind and https://petsc.org/release/faq/
[0]PETSC ERROR: configure using --with-debugging=yes, recompile, link, and run
[0]PETSC ERROR: to get more information on the crash.
[0]PETSC ERROR: ------------------------------------------------------------------------
[0]PETSC ERROR: Caught signal number 13 Broken Pipe: Likely while reading or writing to a socket
[0]PETSC ERROR: Try option -start_in_debugger or -on_error_attach_debugger
[0]PETSC ERROR: or see https://petsc.org/release/faq/#valgrind and https://petsc.org/release/faq/
[0]PETSC ERROR: configure using --with-debugging=yes, recompile, link, and run
[0]PETSC ERROR: to get more information on the crash.
2024-12-02 03:07:48,922 - INFO - Using pre-computed Schur decomposition
Done Hmga2
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_0/model.pt already downloaded
2024-12-02 03:08:53,042 - INFO - Using pre-computed Schur decomposition
Done Setbp1
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_0/model.pt already downloaded
2024-12-02 03:09:57,210 - INFO - Using pre-computed Schur decomposition
Done Cdh1
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_0/model.pt already downloaded
2024-12-02 03:11:01,191 - INFO - Using pre-computed Schur decomposition
Done Hsf2
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_0/model.pt already downloaded
2024-12-02 03:12:04,553 - INFO - Using pre-computed Schur decomposition
Done Rfx6
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_0/model.pt already downloaded
[0]PETSC ERROR: 2024-12-02 03:13:08,114 - INFO - Using pre-computed Schur decomposition
Done Grb10
loading model...
inferring perturbation...
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_1/model.pt already downloaded
2024-12-02 03:14:11,869 - INFO - Using pre-computed Schur decomposition
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_1/model.pt already downloaded
2024-12-02 03:15:15,199 - INFO - Using pre-computed Schur decomposition
Done Rfx3
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_1/model.pt already downloaded
2024-12-02 03:16:18,177 - INFO - Using pre-computed Schur decomposition
Done Adk
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_1/model.pt already downloaded
2024-12-02 03:17:23,089 - INFO - Using pre-computed Schur decomposition
Done Zfp148
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_1/model.pt already downloaded
2024-12-02 03:18:26,791 - INFO - Using pre-computed Schur decomposition
Done Nfatc2
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_1/model.pt already downloaded
2024-12-02 03:19:29,193 - INFO - Using pre-computed Schur decomposition
Done Hmga2
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_1/model.pt already downloaded
2024-12-02 03:20:32,723 - INFO - Using pre-computed Schur decomposition
Done Setbp1
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_1/model.pt already downloaded
2024-12-02 03:21:37,398 - INFO - Using pre-computed Schur decomposition
Done Cdh1
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_1/model.pt already downloaded
2024-12-02 03:22:41,618 - INFO - Using pre-computed Schur decomposition
Done Hsf2
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_1/model.pt already downloaded
[0]PETSC ERROR: ------------------------------------------------------------------------
[0]PETSC ERROR: Caught signal number 13 Broken Pipe: Likely while reading or writing to a socket
[0]PETSC ERROR: Try option -start_in_debugger or -on_error_attach_debugger
[0]PETSC ERROR: or see https://petsc.org/release/faq/#valgrind and https://petsc.org/release/faq/
[0]PETSC ERROR: configure using --with-debugging=yes, recompile, link, and run
[0]PETSC ERROR: to get more information on the crash.
2024-12-02 03:23:45,328 - INFO - Using pre-computed Schur decomposition
Done Rfx6
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_1/model.pt already downloaded
2024-12-02 03:24:48,136 - INFO - Using pre-computed Schur decomposition
Done Grb10
loading model...
inferring perturbation...
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_2/model.pt already downloaded
[0]PETSC ERROR: ------------------------------------------------------------------------
[0]PETSC ERROR: Caught signal number 13 Broken Pipe: Likely while reading or writing to a socket
[0]PETSC ERROR: Try option -start_in_debugger or -on_error_attach_debugger
[0]PETSC ERROR: or see https://petsc.org/release/faq/#valgrind and https://petsc.org/release/faq/
[0]PETSC ERROR: configure using --with-debugging=yes, recompile, link, and run
[0]PETSC ERROR: to get more information on the crash.
2024-12-02 03:25:54,344 - INFO - Using pre-computed Schur decomposition
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_2/model.pt already downloaded
2024-12-02 03:26:59,182 - INFO - Using pre-computed Schur decomposition
Done Rfx3
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_2/model.pt already downloaded
2024-12-02 03:28:03,561 - INFO - Using pre-computed Schur decomposition
Done Adk
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_2/model.pt already downloaded
2024-12-02 03:29:07,961 - INFO - Using pre-computed Schur decomposition
Done Zfp148
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_2/model.pt already downloaded
2024-12-02 03:30:11,971 - INFO - Using pre-computed Schur decomposition
Done Nfatc2
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_2/model.pt already downloaded
2024-12-02 03:31:15,482 - INFO - Using pre-computed Schur decomposition
Done Hmga2
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_2/model.pt already downloaded
2024-12-02 03:32:19,845 - INFO - Using pre-computed Schur decomposition
Done Setbp1
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_2/model.pt already downloaded
2024-12-02 03:33:24,269 - INFO - Using pre-computed Schur decomposition
Done Cdh1
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_2/model.pt already downloaded
[0]PETSC ERROR: ------------------------------------------------------------------------
[0]PETSC ERROR: Caught signal number 13 Broken Pipe: Likely while reading or writing to a socket
[0]PETSC ERROR: Try option -start_in_debugger or -on_error_attach_debugger
[0]PETSC ERROR: or see https://petsc.org/release/faq/#valgrind and https://petsc.org/release/faq/
[0]PETSC ERROR: configure using --with-debugging=yes, recompile, link, and run
[0]PETSC ERROR: to get more information on the crash.
2024-12-02 03:34:28,064 - INFO - Using pre-computed Schur decomposition
Done Hsf2
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_2/model.pt already downloaded
2024-12-02 03:35:31,585 - INFO - Using pre-computed Schur decomposition
Done Rfx6
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_2/model.pt already downloaded
2024-12-02 03:36:35,499 - INFO - Using pre-computed Schur decomposition
Done Grb10
loading model...
inferring perturbation...
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_3/model.pt already downloaded
2024-12-02 03:37:38,550 - INFO - Using pre-computed Schur decomposition
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_3/model.pt already downloaded
2024-12-02 03:38:41,487 - INFO - Using pre-computed Schur decomposition
Done Rfx3
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_3/model.pt already downloaded
2024-12-02 03:39:46,840 - INFO - Using pre-computed Schur decomposition
Done Adk
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_3/model.pt already downloaded
2024-12-02 03:40:49,455 - INFO - Using pre-computed Schur decomposition
Done Zfp148
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_3/model.pt already downloaded
2024-12-02 03:41:54,077 - INFO - Using pre-computed Schur decomposition
Done Nfatc2
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_3/model.pt already downloaded
2024-12-02 03:42:57,801 - INFO - Using pre-computed Schur decomposition
Done Hmga2
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_3/model.pt already downloaded
2024-12-02 03:44:02,697 - INFO - Using pre-computed Schur decomposition
Done Setbp1
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_3/model.pt already downloaded
2024-12-02 03:45:06,043 - INFO - Using pre-computed Schur decomposition
Done Cdh1
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_3/model.pt already downloaded
2024-12-02 03:46:10,795 - INFO - Using pre-computed Schur decomposition
Done Hsf2
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_3/model.pt already downloaded
2024-12-02 03:47:15,665 - INFO - Using pre-computed Schur decomposition
Done Rfx6
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_3/model.pt already downloaded
2024-12-02 03:48:19,237 - INFO - Using pre-computed Schur decomposition
Done Grb10
loading model...
inferring perturbation...
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_4/model.pt already downloaded
[0]PETSC ERROR: ------------------------------------------------------------------------
[0]PETSC ERROR: Caught signal number 13 Broken Pipe: Likely while reading or writing to a socket
[0]PETSC ERROR: Try option -start_in_debugger or -on_error_attach_debugger
[0]PETSC ERROR: or see https://petsc.org/release/faq/#valgrind and https://petsc.org/release/faq/
[0]PETSC ERROR: configure using --with-debugging=yes, recompile, link, and run
[0]PETSC ERROR: to get more information on the crash.
2024-12-02 03:49:24,092 - INFO - Using pre-computed Schur decomposition
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_4/model.pt already downloaded
2024-12-02 03:50:28,964 - INFO - Using pre-computed Schur decomposition
Done Rfx3
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_4/model.pt already downloaded
2024-12-02 03:51:32,437 - INFO - Using pre-computed Schur decomposition
Done Adk
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_4/model.pt already downloaded
2024-12-02 03:52:35,489 - INFO - Using pre-computed Schur decomposition
Done Zfp148
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_4/model.pt already downloaded
2024-12-02 03:53:39,498 - INFO - Using pre-computed Schur decomposition
Done Nfatc2
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_4/model.pt already downloaded
2024-12-02 03:54:43,203 - INFO - Using pre-computed Schur decomposition
Done Hmga2
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_4/model.pt already downloaded
2024-12-02 03:55:47,949 - INFO - Using pre-computed Schur decomposition
Done Setbp1
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_4/model.pt already downloaded
2024-12-02 03:56:52,225 - INFO - Using pre-computed Schur decomposition
Done Cdh1
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_4/model.pt already downloaded
2024-12-02 03:57:56,033 - INFO - Using pre-computed Schur decomposition
Done Hsf2
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_4/model.pt already downloaded
[0]PETSC ERROR: 2024-12-02 03:58:58,973 - INFO - Using pre-computed Schur decomposition
Done Rfx6
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_4/model.pt already downloaded
2024-12-02 04:00:03,972 - INFO - Using pre-computed Schur decomposition
Done Grb10
Plot perturbation results#
Neurod2_target_perturbation = aggregate_model_predictions(
DATA_DIR / DATASET / "results" / "Neurod2_screening_repeat_runs", method="t-statistics"
)
## rank the Epsilon prediction results
with mplscience.style_context(): # Entering the custom style context
sns.set_style("whitegrid")
gene_scores = pd.DataFrame(
{
"Gene": Neurod2_target_perturbation[0].index.tolist(),
"Score": Neurod2_target_perturbation[0].loc[:, "Epsilon"],
}
)
gene_scores.loc[:, "weights"] = gene_scores.loc[:, "Score"].abs()
# gene_scores = gene_scores.sort_values(by='weights', ascending=False).iloc[:10,:]
gene_scores = gene_scores.sort_values(by="Score", ascending=True).iloc[:10, :]
# Create the horizontal bar plot using Seaborn
fig, ax = plt.subplots(figsize=(5, 3))
g = sns.barplot(x="Score", y="Gene", data=gene_scores, color="grey")
# Customize plot aesthetics
g.set_ylabel("Gene", fontsize=14)
g.set_xlabel("Test statistics", fontsize=14)
# Customize tick parameters for better readability
g.tick_params(axis="x", labelsize=14)
g.tick_params(axis="y", labelsize=14)
g.set_title("TF-target regulation perturb screening", fontsize=14)
plt.tight_layout()
plt.setp(g.get_yticklabels(), fontstyle="italic")
if SAVE_FIGURES:
save_path = FIG_DIR / DATASET / "Neurod2_target_screening.svg"
fig.savefig(save_path, format="svg", transparent=True, bbox_inches="tight")
plt.show()