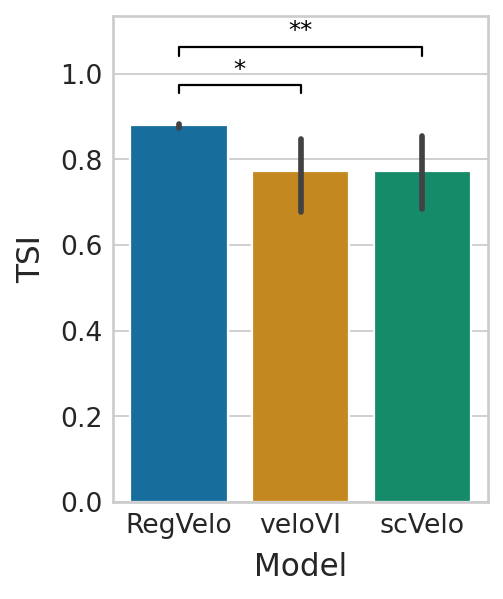
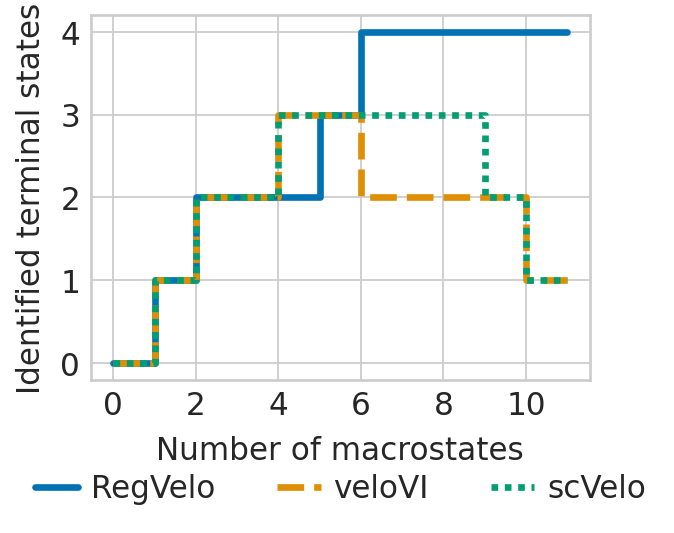
Terminal state identification#
Compares model performance based on terminal state identification.
Library imports#
import numpy as np
import pandas as pd
from scipy.stats import ttest_ind
import matplotlib.pyplot as plt
import mplscience
import seaborn as sns
from matplotlib import rcParams
import cellrank as cr
import scanpy as sc
import scvelo as scv
import scvi
from rgv_tools import DATA_DIR, FIG_DIR
from rgv_tools.benchmarking import get_tsi_score, plot_tsi
from rgv_tools.core import METHOD_PALETTE_TSI
from rgv_tools.plotting._significance import add_significance, get_significance
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_csv from `anndata` is deprecated. Import anndata.io.read_csv instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_loom from `anndata` is deprecated. Import anndata.io.read_loom instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_text from `anndata` is deprecated. Import anndata.io.read_text instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing CSCDataset from `anndata.experimental` is deprecated. Import anndata.abc.CSCDataset instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing CSRDataset from `anndata.experimental` is deprecated. Import anndata.abc.CSRDataset instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_elem from `anndata.experimental` is deprecated. Import anndata.io.read_elem instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_csv from `anndata` is deprecated. Import anndata.io.read_csv instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_excel from `anndata` is deprecated. Import anndata.io.read_excel instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_hdf from `anndata` is deprecated. Import anndata.io.read_hdf instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_loom from `anndata` is deprecated. Import anndata.io.read_loom instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_mtx from `anndata` is deprecated. Import anndata.io.read_mtx instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_text from `anndata` is deprecated. Import anndata.io.read_text instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_umi_tools from `anndata` is deprecated. Import anndata.io.read_umi_tools instead.
warnings.warn(msg, FutureWarning)
General setting#
scvi.settings.seed = 0
[rank: 0] Seed set to 0
plt.rcParams["svg.fonttype"] = "none"
sns.reset_defaults()
sns.reset_orig()
scv.settings.set_figure_params("scvelo", dpi_save=400, dpi=80, transparent=True, fontsize=14, color_map="viridis")
Constants#
DATASET = "zebrafish"
SAVE_FIGURES = False
if SAVE_FIGURES:
(FIG_DIR / DATASET).mkdir(parents=True, exist_ok=True)
FIGURE_FORMATE = "svg"
VELOCITY_METHODS = ["regvelo", "scvelo", "velovi"]
TERMINAL_STATES = [
"mNC_head_mesenchymal",
"mNC_arch2",
"mNC_hox34",
"Pigment",
]
Data loading#
ks = {}
for method in VELOCITY_METHODS:
adata = sc.read_h5ad(DATA_DIR / DATASET / "processed" / f"adata_run_{method}.h5ad")
## construct graph
vk = cr.kernels.VelocityKernel(adata).compute_transition_matrix()
ck = cr.kernels.ConnectivityKernel(adata).compute_transition_matrix()
ks[method] = 0.8 * vk + 0.2 * ck
100%|██████████| 697/697 [00:12<00:00, 55.65cell/s]
100%|██████████| 697/697 [00:00<00:00, 3652.32cell/s]
100%|██████████| 697/697 [00:00<00:00, 3483.05cell/s]
100%|██████████| 697/697 [00:00<00:00, 3748.58cell/s]
100%|██████████| 697/697 [00:00<00:00, 3464.31cell/s]
100%|██████████| 697/697 [00:00<00:00, 3655.55cell/s]
Terminal state identification#
# define threshold from 0.1 to 1
thresholds = np.linspace(0.1, 1, 21)[:20]
estimators = {}
tsi = {}
for method in VELOCITY_METHODS:
estimators[method] = cr.estimators.GPCCA(ks[method])
tsi[method] = get_tsi_score(adata, thresholds, "cell_type", TERMINAL_STATES, estimators[method])
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Requested more macrostates `3` than available Schur vectors `2`. Recomputing the decomposition
WARNING: Requested more macrostates `4` than available Schur vectors `3`. Recomputing the decomposition
WARNING: Requested more macrostates `5` than available Schur vectors `4`. Recomputing the decomposition
WARNING: Requested more macrostates `6` than available Schur vectors `5`. Recomputing the decomposition
WARNING: Requested more macrostates `7` than available Schur vectors `6`. Recomputing the decomposition
WARNING: Requested more macrostates `8` than available Schur vectors `7`. Recomputing the decomposition
WARNING: Requested more macrostates `9` than available Schur vectors `8`. Recomputing the decomposition
WARNING: Requested more macrostates `10` than available Schur vectors `9`. Recomputing the decomposition
WARNING: Requested more macrostates `11` than available Schur vectors `10`. Recomputing the decomposition
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_arch2_2': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_arch2_2': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_arch2_2': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_arch2_2': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_arch2_2': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_arch2_2': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_arch2_2': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_arch2_2': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_arch2_2': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_arch2_2': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_arch2_2': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_arch2_2': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_arch2_2': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_arch2_2': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_arch2_2': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_arch2_2': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_arch2_2': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_arch2_2': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_arch2_2': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_arch2_2': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Requested more macrostates `3` than available Schur vectors `2`. Recomputing the decomposition
WARNING: Requested more macrostates `4` than available Schur vectors `3`. Recomputing the decomposition
WARNING: Requested more macrostates `5` than available Schur vectors `4`. Recomputing the decomposition
WARNING: Requested more macrostates `6` than available Schur vectors `5`. Recomputing the decomposition
WARNING: Requested more macrostates `7` than available Schur vectors `6`. Recomputing the decomposition
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: Requested more macrostates `9` than available Schur vectors `8`. Recomputing the decomposition
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox_1': 28, 'NPB_nohox_2': 28}
WARNING: Requested more macrostates `10` than available Schur vectors `9`. Recomputing the decomposition
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 29, 'NPB_nohox': 25, 'dNC_nohox_2': 23}
WARNING: Requested more macrostates `11` than available Schur vectors `10`. Recomputing the decomposition
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_head_mesenchymal_1': 29, 'mNC_nohox': 27, 'mNC_vagal_1': 27, 'mNC_hox34': 26, 'mNC_head_mesenchymal_2': 26, 'dNC_nohox_2': 23}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox_1': 28, 'NPB_nohox_2': 28}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 29, 'NPB_nohox': 25, 'dNC_nohox_2': 23}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_head_mesenchymal_1': 29, 'mNC_nohox': 27, 'mNC_vagal_1': 27, 'mNC_hox34': 26, 'mNC_head_mesenchymal_2': 26, 'dNC_nohox_2': 23}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox_1': 28, 'NPB_nohox_2': 28}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 29, 'NPB_nohox': 25, 'dNC_nohox_2': 23}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_head_mesenchymal_1': 29, 'mNC_nohox': 27, 'mNC_vagal_1': 27, 'mNC_hox34': 26, 'mNC_head_mesenchymal_2': 26, 'dNC_nohox_2': 23}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox_1': 28, 'NPB_nohox_2': 28}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 29, 'NPB_nohox': 25, 'dNC_nohox_2': 23}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_head_mesenchymal_1': 29, 'mNC_nohox': 27, 'mNC_vagal_1': 27, 'mNC_hox34': 26, 'mNC_head_mesenchymal_2': 26, 'dNC_nohox_2': 23}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox_1': 28, 'NPB_nohox_2': 28}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 29, 'NPB_nohox': 25, 'dNC_nohox_2': 23}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_head_mesenchymal_1': 29, 'mNC_nohox': 27, 'mNC_vagal_1': 27, 'mNC_hox34': 26, 'mNC_head_mesenchymal_2': 26, 'dNC_nohox_2': 23}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox_1': 28, 'NPB_nohox_2': 28}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 29, 'NPB_nohox': 25, 'dNC_nohox_2': 23}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_head_mesenchymal_1': 29, 'mNC_nohox': 27, 'mNC_vagal_1': 27, 'mNC_hox34': 26, 'mNC_head_mesenchymal_2': 26, 'dNC_nohox_2': 23}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox_1': 28, 'NPB_nohox_2': 28}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 29, 'NPB_nohox': 25, 'dNC_nohox_2': 23}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_head_mesenchymal_1': 29, 'mNC_nohox': 27, 'mNC_vagal_1': 27, 'mNC_hox34': 26, 'mNC_head_mesenchymal_2': 26, 'dNC_nohox_2': 23}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox_1': 28, 'NPB_nohox_2': 28}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 29, 'NPB_nohox': 25, 'dNC_nohox_2': 23}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_head_mesenchymal_1': 29, 'mNC_nohox': 27, 'mNC_vagal_1': 27, 'mNC_hox34': 26, 'mNC_head_mesenchymal_2': 26, 'dNC_nohox_2': 23}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox_1': 28, 'NPB_nohox_2': 28}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 29, 'NPB_nohox': 25, 'dNC_nohox_2': 23}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_head_mesenchymal_1': 29, 'mNC_nohox': 27, 'mNC_vagal_1': 27, 'mNC_hox34': 26, 'mNC_head_mesenchymal_2': 26, 'dNC_nohox_2': 23}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox_1': 28, 'NPB_nohox_2': 28}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 29, 'NPB_nohox': 25, 'dNC_nohox_2': 23}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_head_mesenchymal_1': 29, 'mNC_nohox': 27, 'mNC_vagal_1': 27, 'mNC_hox34': 26, 'mNC_head_mesenchymal_2': 26, 'dNC_nohox_2': 23}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox_1': 28, 'NPB_nohox_2': 28}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 29, 'NPB_nohox': 25, 'dNC_nohox_2': 23}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_head_mesenchymal_1': 29, 'mNC_nohox': 27, 'mNC_vagal_1': 27, 'mNC_hox34': 26, 'mNC_head_mesenchymal_2': 26, 'dNC_nohox_2': 23}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox_1': 28, 'NPB_nohox_2': 28}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 29, 'NPB_nohox': 25, 'dNC_nohox_2': 23}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_head_mesenchymal_1': 29, 'mNC_nohox': 27, 'mNC_vagal_1': 27, 'mNC_hox34': 26, 'mNC_head_mesenchymal_2': 26, 'dNC_nohox_2': 23}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox_1': 28, 'NPB_nohox_2': 28}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 29, 'NPB_nohox': 25, 'dNC_nohox_2': 23}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_head_mesenchymal_1': 29, 'mNC_nohox': 27, 'mNC_vagal_1': 27, 'mNC_hox34': 26, 'mNC_head_mesenchymal_2': 26, 'dNC_nohox_2': 23}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox_1': 28, 'NPB_nohox_2': 28}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 29, 'NPB_nohox': 25, 'dNC_nohox_2': 23}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_head_mesenchymal_1': 29, 'mNC_nohox': 27, 'mNC_vagal_1': 27, 'mNC_hox34': 26, 'mNC_head_mesenchymal_2': 26, 'dNC_nohox_2': 23}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox_1': 28, 'NPB_nohox_2': 28}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 29, 'NPB_nohox': 25, 'dNC_nohox_2': 23}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_head_mesenchymal_1': 29, 'mNC_nohox': 27, 'mNC_vagal_1': 27, 'mNC_hox34': 26, 'mNC_head_mesenchymal_2': 26, 'dNC_nohox_2': 23}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox_1': 28, 'NPB_nohox_2': 28}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 29, 'NPB_nohox': 25, 'dNC_nohox_2': 23}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_head_mesenchymal_1': 29, 'mNC_nohox': 27, 'mNC_vagal_1': 27, 'mNC_hox34': 26, 'mNC_head_mesenchymal_2': 26, 'dNC_nohox_2': 23}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox_1': 28, 'NPB_nohox_2': 28}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 29, 'NPB_nohox': 25, 'dNC_nohox_2': 23}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_head_mesenchymal_1': 29, 'mNC_nohox': 27, 'mNC_vagal_1': 27, 'mNC_hox34': 26, 'mNC_head_mesenchymal_2': 26, 'dNC_nohox_2': 23}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox_1': 28, 'NPB_nohox_2': 28}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 29, 'NPB_nohox': 25, 'dNC_nohox_2': 23}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_head_mesenchymal_1': 29, 'mNC_nohox': 27, 'mNC_vagal_1': 27, 'mNC_hox34': 26, 'mNC_head_mesenchymal_2': 26, 'dNC_nohox_2': 23}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox_1': 28, 'NPB_nohox_2': 28}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 29, 'NPB_nohox': 25, 'dNC_nohox_2': 23}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_head_mesenchymal_1': 29, 'mNC_nohox': 27, 'mNC_vagal_1': 27, 'mNC_hox34': 26, 'mNC_head_mesenchymal_2': 26, 'dNC_nohox_2': 23}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox_1': 28, 'NPB_nohox_2': 28}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 29, 'NPB_nohox': 25, 'dNC_nohox_2': 23}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_head_mesenchymal_1': 29, 'mNC_nohox': 27, 'mNC_vagal_1': 27, 'mNC_hox34': 26, 'mNC_head_mesenchymal_2': 26, 'dNC_nohox_2': 23}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Requested more macrostates `3` than available Schur vectors `2`. Recomputing the decomposition
WARNING: Requested more macrostates `4` than available Schur vectors `3`. Recomputing the decomposition
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: Requested more macrostates `6` than available Schur vectors `5`. Recomputing the decomposition
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox': 11}
WARNING: Requested more macrostates `7` than available Schur vectors `6`. Recomputing the decomposition
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: Requested more macrostates `9` than available Schur vectors `8`. Recomputing the decomposition
WARNING: Requested more macrostates `10` than available Schur vectors `9`. Recomputing the decomposition
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox': 29, 'mNC_vagal': 29, 'dNC_hoxa2b': 26}
WARNING: Requested more macrostates `11` than available Schur vectors `10`. Recomputing the decomposition
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox_1': 27, 'mNC_head_mesenchymal_2': 27, 'dNC_hoxa2b_1': 25, 'NPB_nohox': 25, 'mNC_head_mesenchymal_1': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox': 11}
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox': 29, 'mNC_vagal': 29, 'dNC_hoxa2b': 26}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox_1': 27, 'mNC_head_mesenchymal_2': 27, 'dNC_hoxa2b_1': 25, 'NPB_nohox': 25, 'mNC_head_mesenchymal_1': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox': 11}
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox': 29, 'mNC_vagal': 29, 'dNC_hoxa2b': 26}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox_1': 27, 'mNC_head_mesenchymal_2': 27, 'dNC_hoxa2b_1': 25, 'NPB_nohox': 25, 'mNC_head_mesenchymal_1': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox': 11}
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox': 29, 'mNC_vagal': 29, 'dNC_hoxa2b': 26}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox_1': 27, 'mNC_head_mesenchymal_2': 27, 'dNC_hoxa2b_1': 25, 'NPB_nohox': 25, 'mNC_head_mesenchymal_1': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox': 11}
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox': 29, 'mNC_vagal': 29, 'dNC_hoxa2b': 26}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox_1': 27, 'mNC_head_mesenchymal_2': 27, 'dNC_hoxa2b_1': 25, 'NPB_nohox': 25, 'mNC_head_mesenchymal_1': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox': 11}
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox': 29, 'mNC_vagal': 29, 'dNC_hoxa2b': 26}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox_1': 27, 'mNC_head_mesenchymal_2': 27, 'dNC_hoxa2b_1': 25, 'NPB_nohox': 25, 'mNC_head_mesenchymal_1': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox': 11}
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox': 29, 'mNC_vagal': 29, 'dNC_hoxa2b': 26}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox_1': 27, 'mNC_head_mesenchymal_2': 27, 'dNC_hoxa2b_1': 25, 'NPB_nohox': 25, 'mNC_head_mesenchymal_1': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox': 11}
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox': 29, 'mNC_vagal': 29, 'dNC_hoxa2b': 26}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox_1': 27, 'mNC_head_mesenchymal_2': 27, 'dNC_hoxa2b_1': 25, 'NPB_nohox': 25, 'mNC_head_mesenchymal_1': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox': 11}
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox': 29, 'mNC_vagal': 29, 'dNC_hoxa2b': 26}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox_1': 27, 'mNC_head_mesenchymal_2': 27, 'dNC_hoxa2b_1': 25, 'NPB_nohox': 25, 'mNC_head_mesenchymal_1': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox': 11}
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox': 29, 'mNC_vagal': 29, 'dNC_hoxa2b': 26}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox_1': 27, 'mNC_head_mesenchymal_2': 27, 'dNC_hoxa2b_1': 25, 'NPB_nohox': 25, 'mNC_head_mesenchymal_1': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox': 11}
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox': 29, 'mNC_vagal': 29, 'dNC_hoxa2b': 26}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox_1': 27, 'mNC_head_mesenchymal_2': 27, 'dNC_hoxa2b_1': 25, 'NPB_nohox': 25, 'mNC_head_mesenchymal_1': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox': 11}
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox': 29, 'mNC_vagal': 29, 'dNC_hoxa2b': 26}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox_1': 27, 'mNC_head_mesenchymal_2': 27, 'dNC_hoxa2b_1': 25, 'NPB_nohox': 25, 'mNC_head_mesenchymal_1': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox': 11}
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox': 29, 'mNC_vagal': 29, 'dNC_hoxa2b': 26}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox_1': 27, 'mNC_head_mesenchymal_2': 27, 'dNC_hoxa2b_1': 25, 'NPB_nohox': 25, 'mNC_head_mesenchymal_1': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox': 11}
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox': 29, 'mNC_vagal': 29, 'dNC_hoxa2b': 26}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox_1': 27, 'mNC_head_mesenchymal_2': 27, 'dNC_hoxa2b_1': 25, 'NPB_nohox': 25, 'mNC_head_mesenchymal_1': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox': 11}
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox': 29, 'mNC_vagal': 29, 'dNC_hoxa2b': 26}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox_1': 27, 'mNC_head_mesenchymal_2': 27, 'dNC_hoxa2b_1': 25, 'NPB_nohox': 25, 'mNC_head_mesenchymal_1': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox': 11}
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox': 29, 'mNC_vagal': 29, 'dNC_hoxa2b': 26}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox_1': 27, 'mNC_head_mesenchymal_2': 27, 'dNC_hoxa2b_1': 25, 'NPB_nohox': 25, 'mNC_head_mesenchymal_1': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox': 11}
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox': 29, 'mNC_vagal': 29, 'dNC_hoxa2b': 26}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox_1': 27, 'mNC_head_mesenchymal_2': 27, 'dNC_hoxa2b_1': 25, 'NPB_nohox': 25, 'mNC_head_mesenchymal_1': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox': 11}
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox': 29, 'mNC_vagal': 29, 'dNC_hoxa2b': 26}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox_1': 27, 'mNC_head_mesenchymal_2': 27, 'dNC_hoxa2b_1': 25, 'NPB_nohox': 25, 'mNC_head_mesenchymal_1': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox': 11}
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox': 29, 'mNC_vagal': 29, 'dNC_hoxa2b': 26}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox_1': 27, 'mNC_head_mesenchymal_2': 27, 'dNC_hoxa2b_1': 25, 'NPB_nohox': 25, 'mNC_head_mesenchymal_1': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox': 11}
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox': 29, 'mNC_vagal': 29, 'dNC_hoxa2b': 26}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox_1': 27, 'mNC_head_mesenchymal_2': 27, 'dNC_hoxa2b_1': 25, 'NPB_nohox': 25, 'mNC_head_mesenchymal_1': 20}
df = pd.DataFrame(
{
"TSI": tsi["regvelo"] + tsi["velovi"] + tsi["scvelo"],
"Model": ["RegVelo"] * 20 + ["veloVI"] * 20 + ["scVelo"] * 20,
}
)
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(3, 4))
sns.barplot(data=df, x="Model", y="TSI", palette="colorblind", ax=ax)
ttest_res = ttest_ind(tsi["regvelo"], tsi["velovi"], alternative="greater")
significance = get_significance(ttest_res.pvalue)
add_significance(
ax=ax,
left=0,
right=1,
significance=significance,
lw=1,
bracket_level=1.05,
c="k",
level=0,
)
ttest_res = ttest_ind(tsi["regvelo"], tsi["scvelo"], alternative="greater")
significance = get_significance(ttest_res.pvalue)
add_significance(ax=ax, left=0, right=2, significance=significance, lw=1, c="k", level=2, bracket_level=0.9)
y_min, y_max = ax.get_ylim()
ax.set_ylim([y_min, y_max + 0.02])
if SAVE_FIGURES:
fig.savefig(FIG_DIR / DATASET / "tsi_benchmark.svg", format="svg", transparent=True, bbox_inches="tight")
plt.show()
Visualize terminal states#
tsi_rgv_curve = plot_tsi(adata, estimators["regvelo"], 0.8, TERMINAL_STATES, "cell_type")
tsi_scv_curve = plot_tsi(adata, estimators["scvelo"], 0.8, TERMINAL_STATES, "cell_type")
tsi_vi_curve = plot_tsi(adata, estimators["velovi"], 0.8, TERMINAL_STATES, "cell_type")
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_arch2_2': 20}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 26, 'dNC_nohox': 26}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox_1': 28, 'NPB_nohox_2': 28}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hoxa2b': 29, 'NPB_nohox': 25, 'dNC_nohox_2': 23}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'mNC_head_mesenchymal_1': 29, 'mNC_nohox': 27, 'mNC_vagal_1': 27, 'mNC_hox34': 26, 'mNC_head_mesenchymal_2': 26, 'dNC_nohox_2': 23}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox': 11}
WARNING: Unable to compute macrostates with `n_states=7` because it will split complex conjugate eigenvalues. Using `n_states=8`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_hox34': 29}
WARNING: The following terminal states have different number of cells than requested (30): {'NPB_nohox': 29, 'mNC_vagal': 29, 'dNC_hoxa2b': 26}
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'dNC_nohox_1': 27, 'mNC_head_mesenchymal_2': 27, 'dNC_hoxa2b_1': 25, 'NPB_nohox': 25, 'mNC_head_mesenchymal_1': 20}
Plotting#
df = pd.DataFrame(
{
"number_macrostate": range(0, 12),
"RegVelo": [0] + tsi_rgv_curve,
"veloVI": [0] + tsi_vi_curve,
"scVelo": [0] + tsi_scv_curve,
}
)
df = pd.melt(df, ["number_macrostate"])
# Set figure size
with mplscience.style_context():
sns.set_style(style="whitegrid")
rcParams["figure.figsize"] = 4, 3
# Plot the grid plot
ax = sns.lineplot(
x="number_macrostate",
y="value",
hue="variable",
style="variable",
palette=METHOD_PALETTE_TSI,
drawstyle="steps-post",
data=df,
linewidth=3,
)
# Set labels and titles
ax.set(ylabel="Number of correct predictions")
plt.xlabel("Number of macrostates", fontsize=14)
plt.ylabel("Identified terminal states", fontsize=14)
# Customize tick parameters for better readability
ax.set_xticks([0, 2, 4, 6, 8, 10])
ax.set_xticklabels([0, 2, 4, 6, 8, 10])
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.legend(loc="upper center", bbox_to_anchor=(0.5, -0.18), shadow=True, ncol=4, fontsize=14)
if SAVE_FIGURES:
plt.savefig(
FIG_DIR / DATASET / "state_identification_update.svg", format="svg", transparent=True, bbox_inches="tight"
)
plt.show()