Dynamo-based drivers analysis#
Notebook uses dynamo’s LAP analysis to identify key drivers.
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import dynamo as dyn
import scanpy as sc
import scvelo as scv
from dynamo.tools.utils import nearest_neighbors
from rgv_tools import DATA_DIR
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_csv from `anndata` is deprecated. Import anndata.io.read_csv instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_excel from `anndata` is deprecated. Import anndata.io.read_excel instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_hdf from `anndata` is deprecated. Import anndata.io.read_hdf instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_loom from `anndata` is deprecated. Import anndata.io.read_loom instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_mtx from `anndata` is deprecated. Import anndata.io.read_mtx instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_text from `anndata` is deprecated. Import anndata.io.read_text instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_umi_tools from `anndata` is deprecated. Import anndata.io.read_umi_tools instead.
warnings.warn(msg, FutureWarning)
General settings#
plt.rcParams["svg.fonttype"] = "none"
sns.reset_defaults()
sns.reset_orig()
scv.settings.set_figure_params("scvelo", dpi_save=400, dpi=80, transparent=True, fontsize=14, color_map="viridis")
dyn.dynamo_logger.main_silence()
Constants#
DATASET = "hematopoiesis"
SAVE_DATA = True
if SAVE_DATA:
(DATA_DIR / DATASET / "results").mkdir(parents=True, exist_ok=True)
cell_type = ["HSC", "Ery", "Mon"]
fixed_points = np.array(
[
[8.45201833, 9.37697661],
[14.00630381, 2.53853712],
[17.30550636, 6.81561775],
[18.06891717, 11.9840678],
[14.13613403, 15.22244713],
[9.72644402, 14.83745969],
]
)
Data loading#
adata_labeling = sc.read_h5ad(DATA_DIR / DATASET / "raw" / "hsc_dynamo_adata.h5ad")
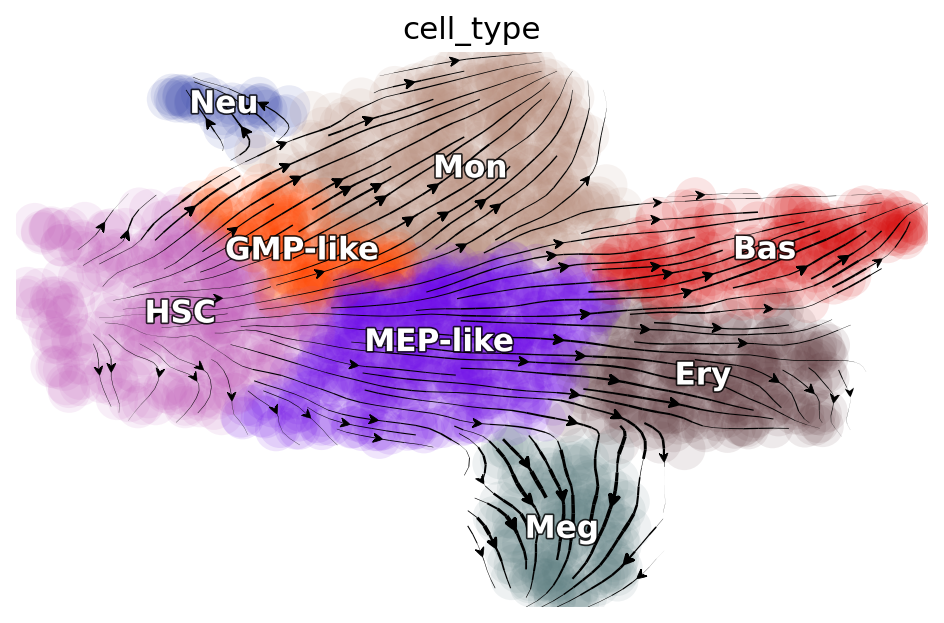
dyn.pl.streamline_plot(adata_labeling, basis="umap", color="cell_type")
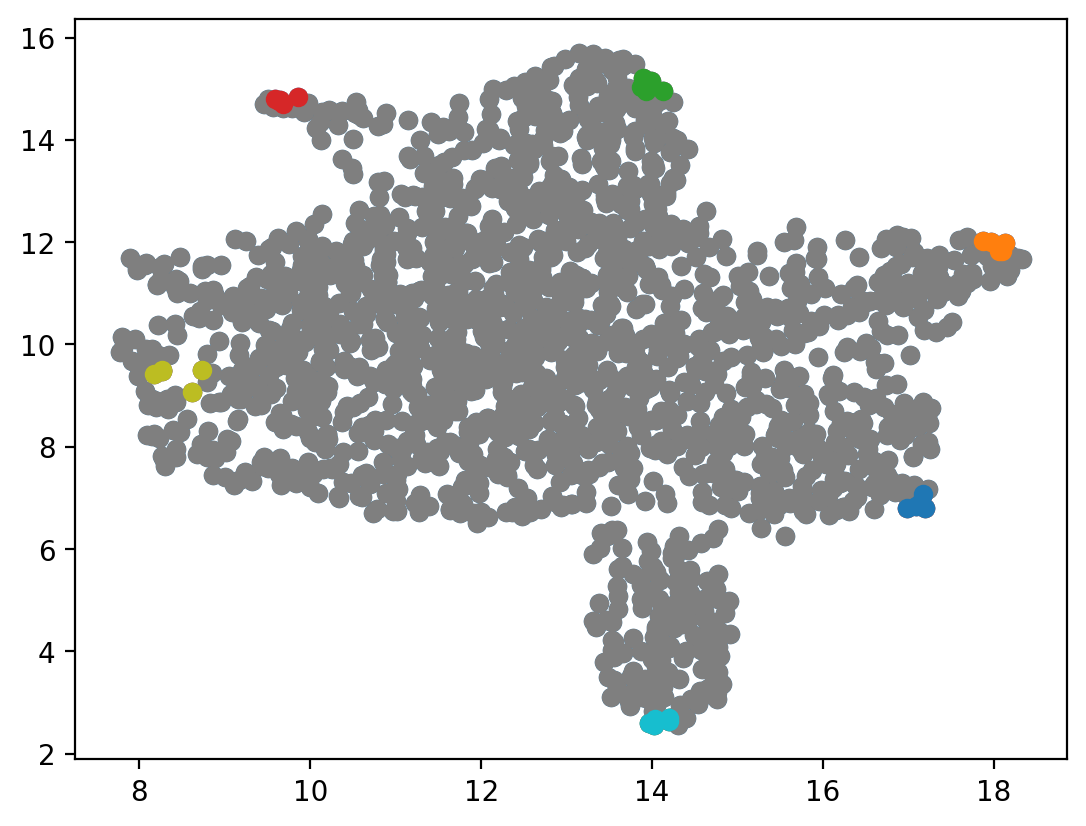
HSC_cells = dyn.tl.select_cell(adata_labeling, "cell_type", "HSC")
Meg_cells = dyn.tl.select_cell(adata_labeling, "cell_type", "Meg")
Ery_cells = dyn.tl.select_cell(adata_labeling, "cell_type", "Ery")
Bas_cells = dyn.tl.select_cell(adata_labeling, "cell_type", "Bas")
Mon_cells = dyn.tl.select_cell(adata_labeling, "cell_type", "Mon")
Neu_cells = dyn.tl.select_cell(adata_labeling, "cell_type", "Neu")
Dynamo pipeline#
HSC_cells_indices = nearest_neighbors(fixed_points[0], adata_labeling.obsm["X_umap"])
Meg_cells_indices = nearest_neighbors(fixed_points[1], adata_labeling.obsm["X_umap"])
Ery_cells_indices = nearest_neighbors(fixed_points[2], adata_labeling.obsm["X_umap"])
Bas_cells_indices = nearest_neighbors(fixed_points[3], adata_labeling.obsm["X_umap"])
Mon_cells_indices = nearest_neighbors(fixed_points[4], adata_labeling.obsm["X_umap"])
Neu_cells_indices = nearest_neighbors(fixed_points[5], adata_labeling.obsm["X_umap"])
plt.scatter(*adata_labeling.obsm["X_umap"].T)
for indices in [
HSC_cells_indices,
Meg_cells_indices,
Ery_cells_indices,
Bas_cells_indices,
Mon_cells_indices,
Neu_cells_indices,
]:
plt.scatter(*adata_labeling[indices[0]].obsm["X_umap"].T)
plt.scatter(*adata_labeling.obsm["X_umap"].T)
for indices in [
HSC_cells_indices,
Meg_cells_indices,
Ery_cells_indices,
Bas_cells_indices,
Mon_cells_indices,
Neu_cells_indices,
]:
plt.scatter(*adata_labeling[indices[0]].obsm["X_umap"].T)
plt.show()
dyn.tl.neighbors(adata_labeling, basis="umap", result_prefix="umap")
|-----> Start computing neighbor graph...
|-----------> X_data is None, fetching or recomputing...
|-----> fetching X data from layer:None, basis:umap
|-----> method arg is None, choosing methods automatically...
|-----------> method kd_tree selected
AnnData object with n_obs × n_vars = 1947 × 21701
obs: 'batch', 'cell_type', 'time', 'nGenes', 'nCounts', 'pMito', 'pass_basic_filter', 'total_Size_Factor', 'initial_total_cell_size', 'spliced_Size_Factor', 'initial_spliced_cell_size', 'Size_Factor', 'initial_cell_size', 'new_Size_Factor', 'initial_new_cell_size', 'unspliced_Size_Factor', 'initial_unspliced_cell_size', 'ntr'
var: 'query', 'scopes', '_id', '_score', 'symbol', 'nCells', 'nCounts', 'pass_basic_filter', 'log_cv', 'score', 'log_m', 'frac', 'use_for_pca'
uns: 'PCs', 'batch_colors', 'cell_type_colors', 'draw_graph', 'dynamics', 'explained_variance_ratio_', 'feature_selection', 'genes_to_use', 'grid_velocity_umap', 'neighbors', 'pca_mean', 'pp', 'velocyto_SVR', 'umap_neighbors'
obsm: 'X_draw_graph_fa', 'X_pca', 'X_umap', 'velocity_umap'
layers: 'M_n', 'M_nn', 'M_s', 'M_ss', 'M_t', 'M_tn', 'M_tt', 'M_u', 'M_us', 'M_uu', 'X_new', 'X_spliced', 'X_total', 'X_unspliced', 'new', 'spliced', 'total', 'unspliced', 'velocity', 'velocity_N', 'velocity_T', 'velocity_alpha_minus_gamma_s'
obsp: 'connectivities', 'cosine_transition_matrix', 'distances', 'moments_con', 'umap_distances', 'umap_connectivities'
dyn.tl.cell_velocities(
adata_labeling,
enforce=True,
X=adata_labeling.layers["M_t"],
V=adata_labeling.layers["velocity_alpha_minus_gamma_s"],
method="cosine",
basis="pca",
)
dyn.vf.VectorField(adata_labeling, basis="pca")
|-----> [calculating transition matrix via cosine kernel with sqrt transform.] in progress: 100.0000%|-----> [calculating transition matrix via cosine kernel with sqrt transform.] completed [206.0417s]
|-----> [projecting velocity vector to low dimensional embedding] in progress: 100.0000%|-----> [projecting velocity vector to low dimensional embedding] completed [0.6028s]
|-----> VectorField reconstruction begins...
|-----> Retrieve X and V based on basis: PCA.
Vector field will be learned in the PCA space.
|-----> Learning vector field with method: sparsevfc.
|-----> [SparseVFC] begins...
|-----> Sampling control points based on data velocity magnitude...
|-----> [SparseVFC] completed [0.3383s]
|-----> [VectorField] completed [0.4921s]
transition_graph = {}
start_cell_indices = [
HSC_cells_indices,
Ery_cells_indices,
Mon_cells_indices,
]
end_cell_indices = start_cell_indices
for i, start in enumerate(start_cell_indices):
for j, end in enumerate(end_cell_indices):
if start is not end:
min_lap_t = True if i == 0 else False
lap = dyn.pd.least_action(
adata_labeling,
[adata_labeling.obs_names[start[0]][0]],
[adata_labeling.obs_names[end[0]][0]],
basis="pca",
adj_key="cosine_transition_matrix",
min_lap_t=min_lap_t,
EM_steps=2,
)
# The `GeneTrajectory` class can be used to output trajectories for any set of genes of interest
gtraj = dyn.pd.GeneTrajectory(adata_labeling)
gtraj.from_pca(lap.X, t=lap.t)
gtraj.calc_msd()
ranking = dyn.vf.rank_genes(adata_labeling, "traj_msd")
print(start, "->", end)
genes = ranking[:5]["all"].to_list()
arr = gtraj.select_gene(genes)
transition_graph[cell_type[i] + "->" + cell_type[j]] = {
"lap": lap,
"LAP_pca": adata_labeling.uns["LAP_pca"],
"ranking": ranking,
"gtraj": gtraj,
}
|-----> searching for the least action path...
|-----> [iterating through 1 pairs] in progress: 100.0000%|-----------> initializing path with the shortest path in the graph built from the velocity transition matrix...
|-----------> optimizing for least action path...
|-----> optimal action: 0.020795
|-----> [iterating through 1 pairs] completed [61.0991s]
|-----> [least action path] completed [61.3269s]
[[1587 1557 1725 1091 1070]] -> [[107 403 22 729 114]]
|-----> searching for the least action path...
|-----> [iterating through 1 pairs] in progress: 100.0000%|-----------> initializing path with the shortest path in the graph built from the velocity transition matrix...
|-----------> optimizing for least action path...
|-----> optimal action: 0.023732
|-----> [iterating through 1 pairs] completed [57.3284s]
|-----> [least action path] completed [57.3649s]
[[1587 1557 1725 1091 1070]] -> [[583 406 82 273 477]]
|-----> searching for the least action path...
|-----> [iterating through 1 pairs] in progress: 100.0000%|-----------> initializing path with the shortest path in the graph built from the velocity transition matrix...
|-----------> optimizing for least action path...
|-----> optimal action: 0.069187
|-----> [iterating through 1 pairs] completed [12.5917s]
|-----> [least action path] completed [12.6136s]
[[107 403 22 729 114]] -> [[1587 1557 1725 1091 1070]]
|-----> searching for the least action path...
|-----> [iterating through 1 pairs] in progress: 100.0000%|-----------> initializing path with the shortest path in the graph built from the velocity transition matrix...
|-----------> optimizing for least action path...
|-----> optimal action: 0.063245
|-----> [iterating through 1 pairs] completed [9.1506s]
|-----> [least action path] completed [9.1867s]
[[107 403 22 729 114]] -> [[583 406 82 273 477]]
|-----> searching for the least action path...
|-----> [iterating through 1 pairs] in progress: 100.0000%|-----------> initializing path with the shortest path in the graph built from the velocity transition matrix...
|-----------> optimizing for least action path...
|-----> optimal action: 0.068055
|-----> [iterating through 1 pairs] completed [10.9457s]
|-----> [least action path] completed [10.9561s]
[[583 406 82 273 477]] -> [[1587 1557 1725 1091 1070]]
|-----> searching for the least action path...
|-----> [iterating through 1 pairs] in progress: 100.0000%|-----------> initializing path with the shortest path in the graph built from the velocity transition matrix...
|-----------> optimizing for least action path...
|-----> optimal action: 0.050282
|-----> [iterating through 1 pairs] completed [7.3260s]
|-----> [least action path] completed [7.3424s]
[[583 406 82 273 477]] -> [[107 403 22 729 114]]
## evaluate ranking from HSC to Mon and Ery
HSC_Mon_ranking = transition_graph["HSC->Mon"]["ranking"]
HSC_Ery_ranking = transition_graph["HSC->Ery"]["ranking"]
Save dataset#
if SAVE_DATA:
HSC_Mon_ranking.to_csv(DATA_DIR / DATASET / "results" / "HSC_Mon_ranking.csv")
HSC_Ery_ranking.to_csv(DATA_DIR / DATASET / "results" / "HSC_Ery_ranking.csv")