Basic preprocessing of the pancreatic endocrine dataset#
Notebook preprocesses the pancreatic endocrine dataset.
Library imports#
import numpy as np
import pandas as pd
import scanpy as sc
import scvelo as scv
from velovi import preprocess_data
from rgv_tools import DATA_DIR
from rgv_tools.preprocessing import set_prior_grn
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_csv from `anndata` is deprecated. Import anndata.io.read_csv instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_excel from `anndata` is deprecated. Import anndata.io.read_excel instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_hdf from `anndata` is deprecated. Import anndata.io.read_hdf instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_loom from `anndata` is deprecated. Import anndata.io.read_loom instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_mtx from `anndata` is deprecated. Import anndata.io.read_mtx instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_text from `anndata` is deprecated. Import anndata.io.read_text instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_umi_tools from `anndata` is deprecated. Import anndata.io.read_umi_tools instead.
warnings.warn(msg, FutureWarning)
General settings#
sc.settings.verbosity = 2
scv.settings.verbosity = 3
Constants#
DATASET = "pancreatic_endocrinogenesis"
(DATA_DIR / DATASET / "raw").mkdir(parents=True, exist_ok=True)
SAVE_DATA = True
if SAVE_DATA:
(DATA_DIR / DATASET / "processed").mkdir(parents=True, exist_ok=True)
Data loading#
adata = scv.datasets.pancreas(file_path=DATA_DIR / DATASET / "raw" / "endocrinogenesis_day15.h5ad")
try downloading from url
https://github.com/theislab/scvelo_notebooks/raw/master/data/Pancreas/endocrinogenesis_day15.h5ad
... this may take a while but only happens once
creating directory data/Pancreas/ for saving data
TF = pd.read_csv(DATA_DIR / DATASET / "raw" / "allTFs_mm.txt", header=None)
gt_net = pd.read_csv(DATA_DIR / DATASET / "raw" / "skeleton.csv", index_col=0)
Preprocessing#
scv.pp.filter_and_normalize(adata, min_shared_counts=10, n_top_genes=2000)
Filtered out 19641 genes that are detected 10 counts (shared).
Normalized count data: X, spliced, unspliced.
Extracted 2000 highly variable genes.
Logarithmized X.
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/scvelo/preprocessing/utils.py:705: DeprecationWarning: `log1p` is deprecated since scVelo v0.3.0 and will be removed in a future version. Please use `log1p` from `scanpy.pp` instead.
log1p(adata)
sc.pp.neighbors(adata, n_neighbors=30, n_pcs=30)
computing neighbors
using 'X_pca' with n_pcs = 30
finished (0:00:23)
scv.pp.moments(adata, n_pcs=None, n_neighbors=None)
adata
computing moments based on connectivities
finished (0:00:00) --> added
'Ms' and 'Mu', moments of un/spliced abundances (adata.layers)
AnnData object with n_obs × n_vars = 3696 × 2000
obs: 'clusters_coarse', 'clusters', 'S_score', 'G2M_score', 'initial_size_unspliced', 'initial_size_spliced', 'initial_size', 'n_counts'
var: 'highly_variable_genes', 'gene_count_corr', 'means', 'dispersions', 'dispersions_norm', 'highly_variable'
uns: 'clusters_coarse_colors', 'clusters_colors', 'day_colors', 'neighbors', 'pca', 'log1p'
obsm: 'X_pca', 'X_umap'
layers: 'spliced', 'unspliced', 'Ms', 'Mu'
obsp: 'distances', 'connectivities'
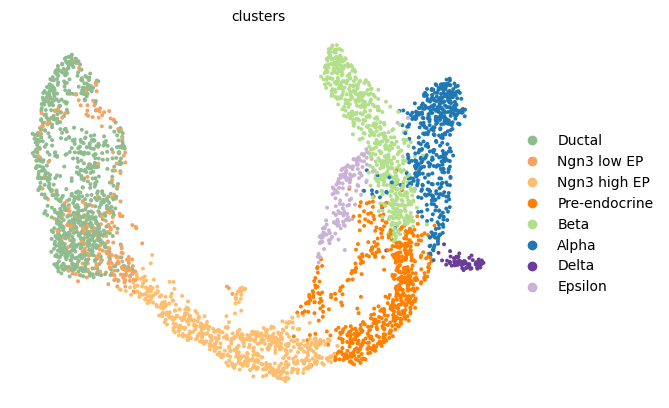
scv.pl.umap(adata, color="clusters", legend_loc="right")
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/scvelo/plotting/utils.py:63: DeprecationWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, pd.CategoricalDtype) instead
return isinstance(c, str) and c in data.obs.keys() and cat(data.obs[c])
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/IPython/core/pylabtools.py:77: DeprecationWarning: backend2gui is deprecated since IPython 8.24, backends are managed in matplotlib and can be externally registered.
warnings.warn(
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/scvelo/plotting/utils.py:63: DeprecationWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, pd.CategoricalDtype) instead
return isinstance(c, str) and c in data.obs.keys() and cat(data.obs[c])
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/scvelo/plotting/utils.py:63: DeprecationWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, pd.CategoricalDtype) instead
return isinstance(c, str) and c in data.obs.keys() and cat(data.obs[c])
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/scvelo/plotting/utils.py:63: DeprecationWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, pd.CategoricalDtype) instead
return isinstance(c, str) and c in data.obs.keys() and cat(data.obs[c])
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/scvelo/plotting/utils.py:63: DeprecationWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, pd.CategoricalDtype) instead
return isinstance(c, str) and c in data.obs.keys() and cat(data.obs[c])
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/scvelo/plotting/utils.py:63: DeprecationWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, pd.CategoricalDtype) instead
return isinstance(c, str) and c in data.obs.keys() and cat(data.obs[c])
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/scvelo/plotting/utils.py:63: DeprecationWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, pd.CategoricalDtype) instead
return isinstance(c, str) and c in data.obs.keys() and cat(data.obs[c])
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/scvelo/plotting/utils.py:63: DeprecationWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, pd.CategoricalDtype) instead
return isinstance(c, str) and c in data.obs.keys() and cat(data.obs[c])
RegVelo preprocessing#
adata = set_prior_grn(adata, gt_net, keep_dim=True) ## keep_dim = True due to the the sparse prior GRN
velocity_genes = preprocess_data(adata.copy()).var_names.tolist()
computing velocities
finished (0:00:00) --> added
'velocity', velocity vectors for each individual cell (adata.layers)
tf_grn = adata.var_names[adata.uns["skeleton"].T.sum(0) != 0].tolist()
tfs = list(set(TF.iloc[:, 0].tolist()).intersection(tf_grn))
adata.var["tf"] = adata.var_names.isin(tfs)
var_mask = np.union1d(adata.var_names[adata.var["tf"]], velocity_genes)
adata = adata[:, var_mask].copy()
adata = preprocess_data(adata, filter_on_r2=False)
# Filter the skeleton matrix `W` based on the selected indices
skeleton = adata.uns["skeleton"].loc[adata.var_names.tolist(), adata.var_names.tolist()]
# Update the filtered values in `uns`
adata.uns.update({"skeleton": skeleton, "regulators": adata.var_names.tolist(), "targets": adata.var_names.tolist()})
## focus on velocity genes to ensure calculation stability of scvelo and veloVI
adata.var["velocity_genes"] = adata.var_names.isin(velocity_genes)
Save dataset#
if SAVE_DATA:
adata.write_h5ad(DATA_DIR / DATASET / "processed" / "adata_preprocessed.h5ad")