Visualize GRN#
Notebook visualize regvelo-inferred cell cycle core GRN
Library imports#
import networkx as nx
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import anndata as ad
import scvelo as scv
import scvi
from regvelo import REGVELOVI
from rgv_tools import DATA_DIR, FIG_DIR
from rgv_tools.perturbation import inferred_GRN
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_csv from `anndata` is deprecated. Import anndata.io.read_csv instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_excel from `anndata` is deprecated. Import anndata.io.read_excel instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_hdf from `anndata` is deprecated. Import anndata.io.read_hdf instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_loom from `anndata` is deprecated. Import anndata.io.read_loom instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_mtx from `anndata` is deprecated. Import anndata.io.read_mtx instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_text from `anndata` is deprecated. Import anndata.io.read_text instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_umi_tools from `anndata` is deprecated. Import anndata.io.read_umi_tools instead.
warnings.warn(msg, FutureWarning)
2024-12-12 15:01:18.961132: E external/local_xla/xla/stream_executor/cuda/cuda_fft.cc:477] Unable to register cuFFT factory: Attempting to register factory for plugin cuFFT when one has already been registered
WARNING: All log messages before absl::InitializeLog() is called are written to STDERR
E0000 00:00:1734012078.978424 1042103 cuda_dnn.cc:8310] Unable to register cuDNN factory: Attempting to register factory for plugin cuDNN when one has already been registered
E0000 00:00:1734012078.983920 1042103 cuda_blas.cc:1418] Unable to register cuBLAS factory: Attempting to register factory for plugin cuBLAS when one has already been registered
General settings#
%matplotlib inline
plt.rcParams["svg.fonttype"] = "none"
sns.reset_defaults()
sns.reset_orig()
scv.settings.set_figure_params("scvelo", dpi_save=400, dpi=80, transparent=True, fontsize=14, color_map="viridis")
scvi.settings.seed = 0
[rank: 0] Seed set to 0
Constants#
DATASET = "cell_cycle"
SAVE_DATA = True
SAVE_FIGURES = True
if SAVE_DATA:
(DATA_DIR / DATASET / "results").mkdir(parents=True, exist_ok=True)
if SAVE_FIGURES:
(FIG_DIR / DATASET / "results").mkdir(parents=True, exist_ok=True)
Data loading#
adata = ad.io.read_h5ad(DATA_DIR / DATASET / "processed" / "adata_processed.h5ad")
TF = pd.read_csv(DATA_DIR / DATASET / "raw" / "allTFs_hg38.txt", header=None)
TF = adata.var_names[adata.var_names.isin(TF.iloc[:, 0])]
adata
AnnData object with n_obs × n_vars = 1146 × 395
obs: 'phase', 'fucci_time', 'initial_size_unspliced', 'initial_size_spliced', 'initial_size', 'n_counts'
var: 'ensum_id', 'gene_count_corr', 'means', 'dispersions', 'dispersions_norm', 'highly_variable', 'velocity_gamma', 'velocity_qreg_ratio', 'velocity_r2', 'velocity_genes'
uns: 'log1p', 'neighbors', 'pca', 'umap', 'velocity_params'
obsm: 'X_pca', 'X_umap'
varm: 'PCs', 'true_skeleton'
layers: 'Ms', 'Mu', 'spliced', 'total', 'unspliced', 'velocity'
obsp: 'connectivities', 'distances'
Model loading#
vae = REGVELOVI.load(DATA_DIR / DATASET / "regvelo_model", adata)
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/cell_cycle/regvelo_model/model.pt already
downloaded
Identify most activate TF#
adata.obs["cluster"] = "0" ## create label with one-class because of it is cell line dataset
GRN = inferred_GRN(vae, adata, label="cluster", group="all")
pd.DataFrame((GRN[:, adata.var.index.isin(TF)]).mean(0), index=adata.var.index[adata.var.index.isin(TF)]).sort_values(
0, ascending=False
).iloc[:10, :]
Computing global GRN...
| 0 | |
|---|---|
| TGIF1 | 1.025551 |
| ETV7 | 0.316861 |
| ETV1 | 0.300762 |
| KLF7 | 0.192435 |
| ARID5B | 0.153252 |
| EGR1 | 0.140843 |
| KIF22 | 0.044913 |
| EBF1 | 0.018076 |
| SMAP2 | -0.000480 |
| ZSCAN4 | -0.024477 |
GRN processing pipeline#
## Detect regulon for TGIF1
targets = GRN[:, [i == "TGIF1" for i in adata.var.index]].reshape(-1)
targets = pd.DataFrame(targets, index=adata.var.index)
targets.loc[:, "weight"] = targets.iloc[:, 0].abs()
GRN_visualize_tgif1 = targets.sort_values("weight", ascending=False).iloc[:50, :]
## Detect regulon for ETV1
targets = GRN[:, [i == "ETV1" for i in adata.var.index]].reshape(-1)
targets = pd.DataFrame(targets, index=adata.var.index)
targets.loc[:, "weight"] = targets.iloc[:, 0].abs()
GRN_visualize_etv1 = targets.sort_values("weight", ascending=False).iloc[:50, :]
## processing GRN
df1 = pd.DataFrame({"from": ["TGIF1"] * 50, "to": GRN_visualize_tgif1.index.tolist()})
df2 = pd.DataFrame({"from": ["ETV1"] * 50, "to": GRN_visualize_etv1.index.tolist()})
df = pd.concat([df1, df2], axis=0)
list1 = ["TGIF1"] + GRN_visualize_tgif1.index.tolist()
list2 = ["ETV1"] + GRN_visualize_etv1.index.tolist()
# Define communities
community1 = list(set(list1) - set(list2)) # Nodes only in list1
community2 = list(set(list2) - set(list1)) # Nodes only in list2
community3 = list(set(list1).intersection(list2)) # Nodes shared by both
communities = [frozenset(set(community1)), frozenset(set(community2)), frozenset(set(community3))]
# return GRN
G = nx.from_pandas_edgelist(df, source="from", target="to", create_using=nx.DiGraph())
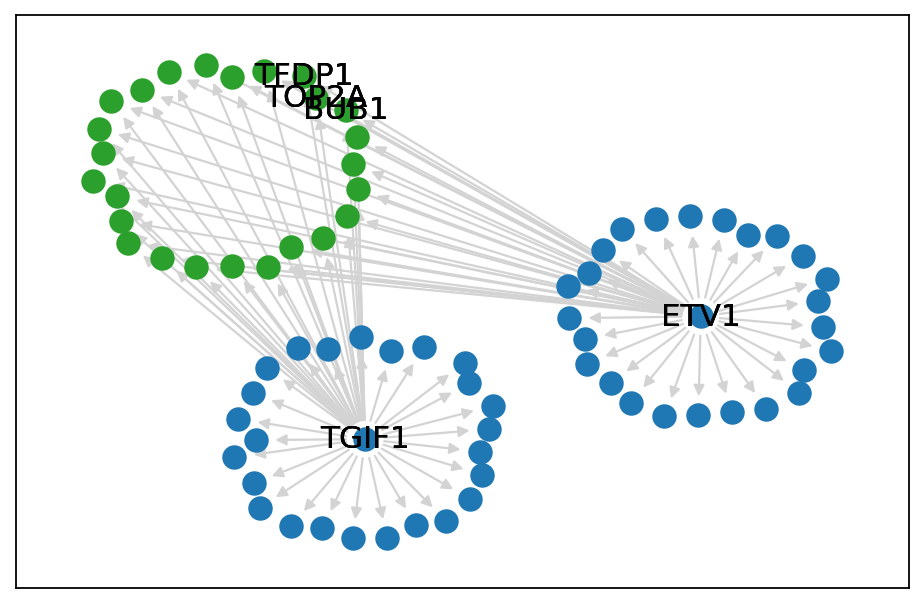
GRN visualization#
if SAVE_FIGURES:
fig, ax = plt.subplots(figsize=(6, 4))
supergraph = nx.cycle_graph(len(communities))
superpos = nx.spring_layout(G, scale=3)
# Use the "supernode" positions as the center of each node cluster
centers = list(superpos.values())
pos = {}
for center, comm in zip(centers, communities):
pos.update(nx.spring_layout(nx.subgraph(G, comm), center=center))
# Nodes colored by cluster
node_list = ["TGIF1", "ETV1", "BUB1", "TOP2A", "TFDP1"]
for nodes, clr in zip(communities, ("tab:blue", "tab:blue", "tab:green")):
nx.draw_networkx_nodes(G, pos=pos, nodelist=nodes, node_color=clr, node_size=100)
nx.draw_networkx_labels(G, pos=pos, labels={node: node for node in node_list}, font_size=14, font_color="black")
nx.draw_networkx_edges(G, pos=pos, edge_color="lightgrey")
plt.tight_layout()
fig.savefig(FIG_DIR / DATASET / "O2SC_GRN.svg", format="svg", transparent=True, bbox_inches="tight")
plt.show()