Comparing uncertainty#
Notebook compares the uncertainty estimates of RegVelo and VeloVI.
Library imports#
import numpy as np
import pandas as pd
from scipy.stats import wilcoxon
import matplotlib.pyplot as plt
import mplscience
import seaborn as sns
import cellrank as cr
import scanpy as sc
import scvelo as scv
from rgv_tools import DATA_DIR, FIG_DIR
from rgv_tools.core import SIGNIFICANCE_PALETTE
from rgv_tools.plotting._significance import get_significance
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_csv from `anndata` is deprecated. Import anndata.io.read_csv instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_excel from `anndata` is deprecated. Import anndata.io.read_excel instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_hdf from `anndata` is deprecated. Import anndata.io.read_hdf instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_loom from `anndata` is deprecated. Import anndata.io.read_loom instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_mtx from `anndata` is deprecated. Import anndata.io.read_mtx instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_text from `anndata` is deprecated. Import anndata.io.read_text instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_umi_tools from `anndata` is deprecated. Import anndata.io.read_umi_tools instead.
warnings.warn(msg, FutureWarning)
General settings#
plt.rcParams["svg.fonttype"] = "none"
scv.settings.set_figure_params("scvelo", dpi_save=400, dpi=80, transparent=True, fontsize=14, color_map="viridis")
Constants#
DATASET = "hematopoiesis"
SAVE_FIGURES = False
if SAVE_FIGURES:
(FIG_DIR / DATASET).mkdir(parents=True, exist_ok=True)
FIGURE_FORMATE = "svg"
terminal_states = ["Meg", "Mon", "Bas", "Ery", "Neu"]
Data loading#
uncertainty_rgv = pd.read_csv(DATA_DIR / DATASET / "results" / "uncertainty_rgv.csv")
uncertainty_vi = pd.read_csv(DATA_DIR / DATASET / "results" / "uncertainty_vi.csv")
adata = sc.read_h5ad(DATA_DIR / DATASET / "processed" / "adata_run_regvelo.h5ad")
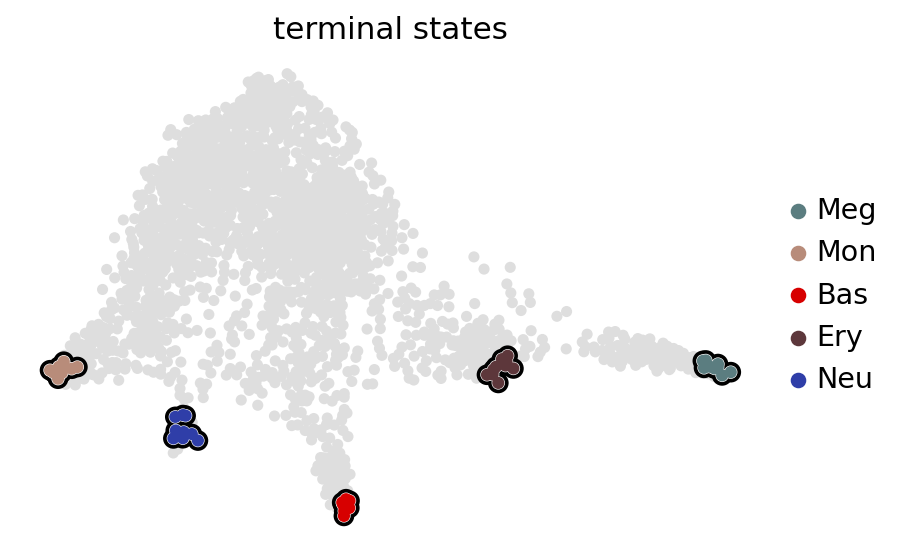
Define terminal states#
vk = cr.kernels.VelocityKernel(adata)
vk.compute_transition_matrix()
ck = cr.kernels.ConnectivityKernel(adata).compute_transition_matrix()
estimator = cr.estimators.GPCCA(0.8 * vk + 0.2 * ck)
## evaluate the fate prob on original space
estimator.compute_macrostates(n_states=6, cluster_key="cell_type", n_cells=10)
estimator.set_terminal_states(terminal_states, n_cells=10)
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
GPCCA[kernel=(0.8 * VelocityKernel[n=1947] + 0.2 * ConnectivityKernel[n=1947]), initial_states=None, terminal_states=['Bas', 'Ery', 'Meg', 'Mon', 'Neu']]
estimator.plot_macrostates(which="terminal", basis="draw_graph_fa", legend_loc="right", s=100)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/scvelo/plotting/scatter.py:656: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
smp = ax.scatter(
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/scvelo/plotting/scatter.py:694: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
ax.scatter(
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/scvelo/plotting/utils.py:1391: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
ax.scatter(x, y, s=bg_size, marker=".", c=bg_color, zorder=zord - 2, **kwargs)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/scvelo/plotting/utils.py:1392: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
ax.scatter(x, y, s=gp_size, marker=".", c=gp_color, zorder=zord - 1, **kwargs)
uncertainty_rgv["terminal_state"] = estimator._term_states[0].tolist()
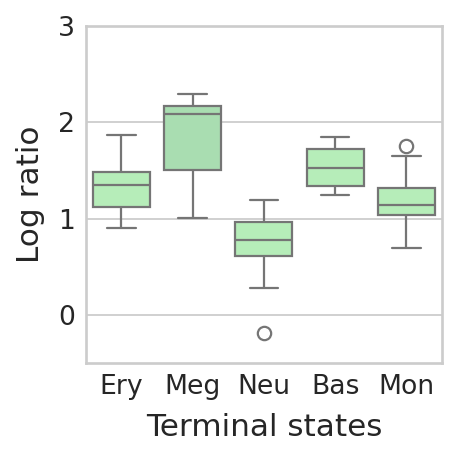
Compare the ratio of each terminal state’s uncertainty score to the mean value of the progenitor cells#
regvi_score = uncertainty_rgv["uncertainty"].values / np.mean(
uncertainty_rgv["uncertainty"].values[adata.obs["cell_type"].isin(["MEP-like", "GMP-like"])]
)
vi_score = uncertainty_vi["uncertainty"].values / np.mean(
uncertainty_vi["uncertainty"].values[adata.obs["cell_type"].isin(["MEP-like", "GMP-like"])]
)
terminal_state = uncertainty_rgv["terminal_state"]
palette = []
for ct in terminal_state.unique()[1:].tolist():
data = np.log10(vi_score[terminal_state == ct] / regvi_score[terminal_state == ct])
# Step 2: Perform a one-sample t-test to compare the mean to a threshold value (e.g., 5)
threshold = 0
b = np.zeros(len(data))
t_stat, p_value = wilcoxon(data, b, alternative="greater")
significance = get_significance(p_value)
palette.append(SIGNIFICANCE_PALETTE[significance])
scores = {
"Ery": np.log10(vi_score[terminal_state == "Ery"] / regvi_score[terminal_state == "Ery"]),
"Meg": np.log10(vi_score[terminal_state == "Meg"] / regvi_score[terminal_state == "Meg"]),
"Neu": np.log10(vi_score[terminal_state == "Neu"] / regvi_score[terminal_state == "Neu"]),
"Bas": np.log10(vi_score[terminal_state == "Bas"] / regvi_score[terminal_state == "Bas"]),
"Mon": np.log10(vi_score[terminal_state == "Mon"] / regvi_score[terminal_state == "Mon"]),
}
# Combine into a DataFrame
df = pd.DataFrame(scores)
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, axes = plt.subplots(figsize=(3, 3))
# Step 4: Color based on significance
sns.boxplot(data=df, palette=palette)
# Add titles and labels
plt.ylim(-0.5, 3)
plt.xlabel("Terminal states")
plt.ylabel("Log ratio")
fig.tight_layout()
if SAVE_FIGURES:
fig.savefig(
FIG_DIR / DATASET / "terminal_states_noise_ratio.svg", format="svg", transparent=True, bbox_inches="tight"
)
plt.show()