Performance comparison of inference on hematopoiesis dataset#
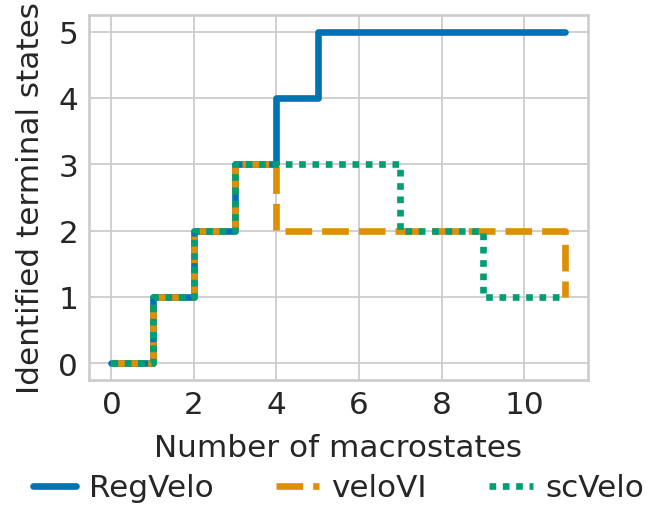
Comparison of cross-boundary correctness (CBC) and terminal state identification
Library imports#
from tqdm import tqdm
import numpy as np
import pandas as pd
from scipy.stats import ttest_ind
import matplotlib.pyplot as plt
import mplscience
import seaborn as sns
from matplotlib import rcParams
import cellrank as cr
import scanpy as sc
import scvelo as scv
from rgv_tools import DATA_DIR, FIG_DIR
from rgv_tools.benchmarking import get_tsi_score, plot_tsi
from rgv_tools.core import METHOD_PALETTE_TSI
from rgv_tools.plotting._significance import add_significance, get_significance
from rgv_tools.utils._stools import reverse_cluster, reverse_cluster_dict
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_csv from `anndata` is deprecated. Import anndata.io.read_csv instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_excel from `anndata` is deprecated. Import anndata.io.read_excel instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_hdf from `anndata` is deprecated. Import anndata.io.read_hdf instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_loom from `anndata` is deprecated. Import anndata.io.read_loom instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_mtx from `anndata` is deprecated. Import anndata.io.read_mtx instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_text from `anndata` is deprecated. Import anndata.io.read_text instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_umi_tools from `anndata` is deprecated. Import anndata.io.read_umi_tools instead.
warnings.warn(msg, FutureWarning)
General settings#
plt.rcParams["svg.fonttype"] = "none"
sns.reset_defaults()
sns.reset_orig()
scv.settings.set_figure_params("scvelo", dpi_save=400, dpi=80, transparent=True, fontsize=14, color_map="viridis")
Constants#
DATASET = "hematopoiesis"
SAVE_FIGURES = False
if SAVE_FIGURES:
(FIG_DIR / DATASET).mkdir(parents=True, exist_ok=True)
FIGURE_FORMATE = "svg"
VELOCITY_METHODS = ["regvelo_unregularized", "regvelo", "scvelo", "velovi"]
TERMINAL_STATES = ["Ery", "Mon", "Bas", "Neu", "Meg"]
STATE_TRANSITIONS = [
("HSC", "GMP-like"),
("GMP-like", "Mon"),
("HSC", "MEP-like"),
("MEP-like", "Ery"),
("MEP-like", "Meg"),
("MEP-like", "Bas"),
]
STATE_TRANSITIONS_REVERSE = [
("GMP-like", "HSC"),
("Mon", "GMP-like"),
("MEP-like", "HSC"),
("Ery", "MEP-like"),
("Meg", "MEP-like"),
("Bas", "MEP-like"),
]
Data loading#
vks = {}
for method in VELOCITY_METHODS:
adata = sc.read_h5ad(DATA_DIR / DATASET / "processed" / f"adata_run_{method}.h5ad")
del adata.var["velocity_genes"]
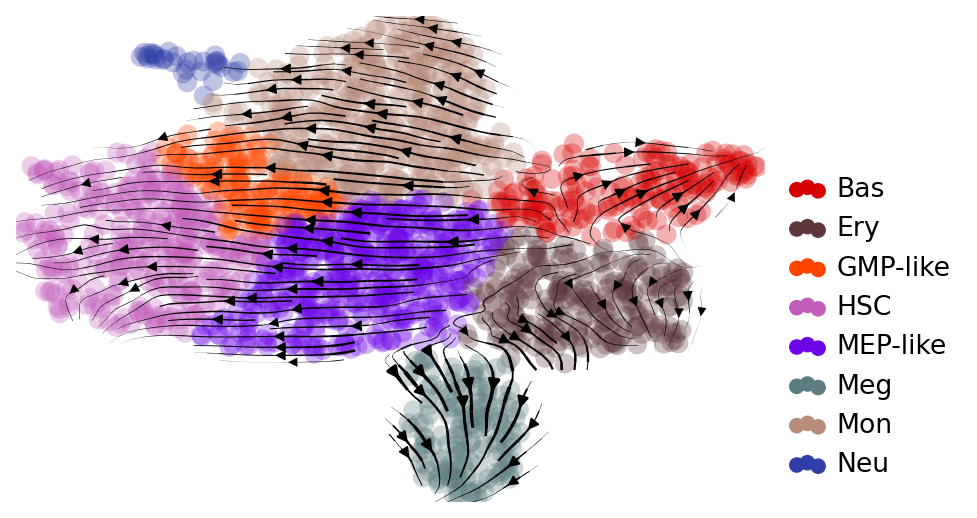
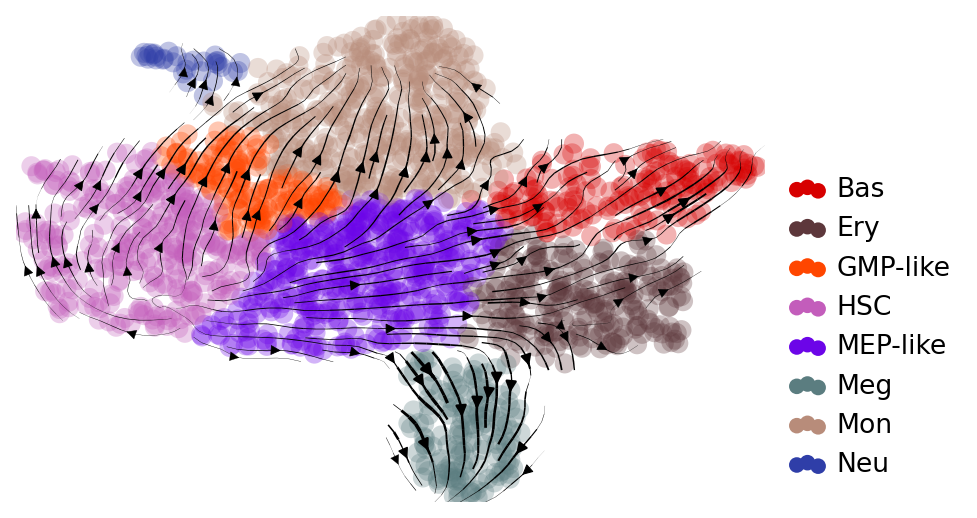
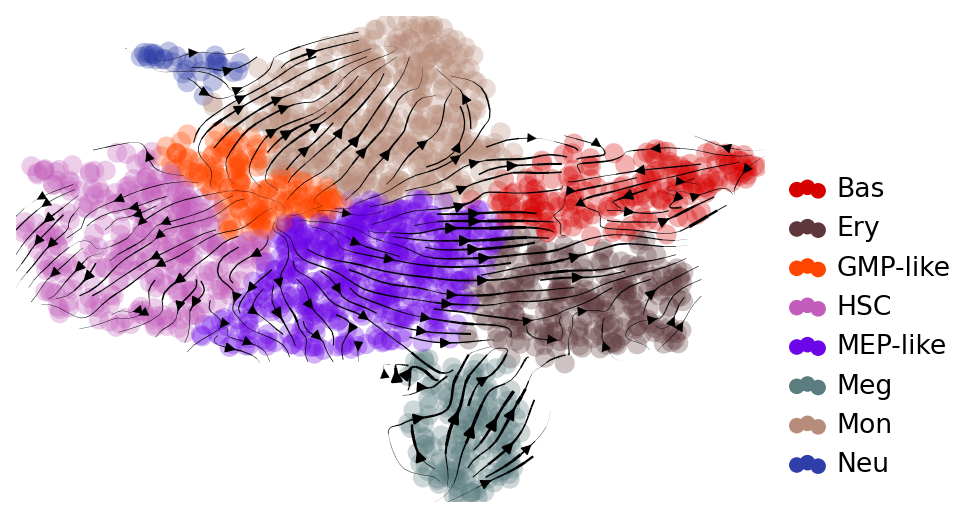
## Visualize velocity on UMAP
with mplscience.style_context():
fig, ax = plt.subplots(figsize=(6, 4))
scv.tl.velocity_graph(adata)
scv.pl.velocity_embedding_stream(
adata, basis="umap", color="cell_type", title="", legend_loc="lower right", ax=ax
)
if SAVE_FIGURES:
fig.savefig(
FIG_DIR / DATASET / f"vector_field_{method}.svg", format="svg", transparent=True, bbox_inches="tight"
)
plt.show()
## construct graph
vk = cr.kernels.VelocityKernel(adata)
vk.compute_transition_matrix()
vks[method] = vk
computing velocity graph (using 1/128 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:00) --> added
'velocity_umap', embedded velocity vectors (adata.obsm)
computing velocity graph (using 1/128 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:00) --> added
'velocity_umap', embedded velocity vectors (adata.obsm)
computing velocity graph (using 1/128 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:00) --> added
'velocity_umap', embedded velocity vectors (adata.obsm)
computing velocity graph (using 1/128 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:00) --> added
'velocity_umap', embedded velocity vectors (adata.obsm)
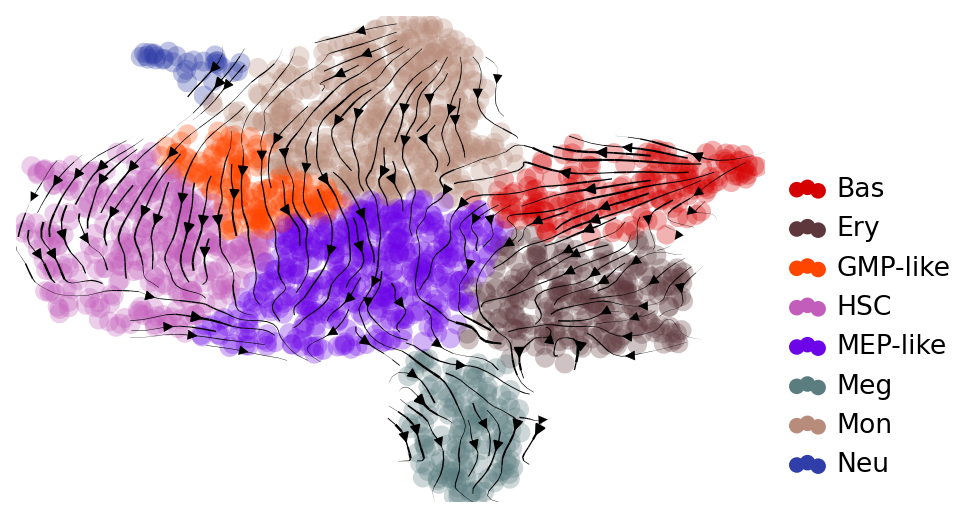
Comparing RegVelo and scVelo#
Measuring forward CBC#
cluster_key = "cell_type"
rep = "X_pca"
score_df = []
for source, target in tqdm(STATE_TRANSITIONS):
cbc_rgv = vks["regvelo"].cbc(source=source, target=target, cluster_key=cluster_key, rep=rep)
cbc_rgv_wo = vks["regvelo_unregularized"].cbc(source=source, target=target, cluster_key=cluster_key, rep=rep)
score_df.append(
pd.DataFrame(
{
"State transition": [f"{source} - {target}"] * len(cbc_rgv),
"Log ratio": np.log((cbc_rgv + 1) / (cbc_rgv_wo + 1)),
}
)
)
score_df_forward = pd.concat(score_df)
100%|██████████| 6/6 [00:01<00:00, 5.96it/s]
dfs = []
ttest_res = {}
significances = {}
for source, target in STATE_TRANSITIONS:
obs_mask = score_df_forward["State transition"].isin([f"{source} - {target}"])
a = score_df_forward.loc[obs_mask, "Log ratio"].values
b = np.zeros(len(a))
ttest_res[f"{source} - {target}"] = ttest_ind(a, b, equal_var=False, alternative="greater")
significances[f"{source} - {target}"] = get_significance(ttest_res[f"{source} - {target}"].pvalue)
significances_forward = significances.copy()
significances_forward
{'HSC - GMP-like': '***',
'GMP-like - Mon': '***',
'HSC - MEP-like': '***',
'MEP-like - Ery': '***',
'MEP-like - Meg': 'n.s.',
'MEP-like - Bas': '***'}
Measuring backward CBC#
score_df = []
for source, target in tqdm(STATE_TRANSITIONS_REVERSE):
cbc_rgv = -vks["regvelo"].cbc(source=source, target=target, cluster_key=cluster_key, rep=rep)
cbc_rgv_wo = -vks["regvelo_unregularized"].cbc(source=source, target=target, cluster_key=cluster_key, rep=rep)
score_df.append(
pd.DataFrame(
{
"State transition": [f"{source} - {target}"] * len(cbc_rgv),
"Log ratio": np.log((cbc_rgv + 1) / (cbc_rgv_wo + 1)),
}
)
)
score_df_backward = pd.concat(score_df)
100%|██████████| 6/6 [00:00<00:00, 6.58it/s]
dfs = []
ttest_res = {}
significances = {}
for source, target in STATE_TRANSITIONS_REVERSE:
obs_mask = score_df_backward["State transition"].isin([f"{source} - {target}"])
a = score_df_backward.loc[obs_mask, "Log ratio"].values
b = np.zeros(len(a))
ttest_res[f"{source} - {target}"] = ttest_ind(a, b, equal_var=False, alternative="greater")
significances[f"{source} - {target}"] = get_significance(ttest_res[f"{source} - {target}"].pvalue)
significances_backward = significances.copy()
significances_backward
{'GMP-like - HSC': '***',
'Mon - GMP-like': '***',
'MEP-like - HSC': '***',
'Ery - MEP-like': '***',
'Meg - MEP-like': '*',
'Bas - MEP-like': 'n.s.'}
score_df_backward["State transition"] = score_df_backward["State transition"].apply(reverse_cluster)
score_df_forward.loc[:, "Direction"] = "Forward"
score_df_backward.loc[:, "Direction"] = "Backward"
score_df = pd.concat([score_df_forward, score_df_backward])
significances_backward = reverse_cluster_dict(significances_backward)
significances_backward
{'HSC - GMP-like': '***',
'GMP-like - Mon': '***',
'HSC - MEP-like': '***',
'MEP-like - Ery': '***',
'MEP-like - Meg': '*',
'MEP-like - Bas': 'n.s.'}
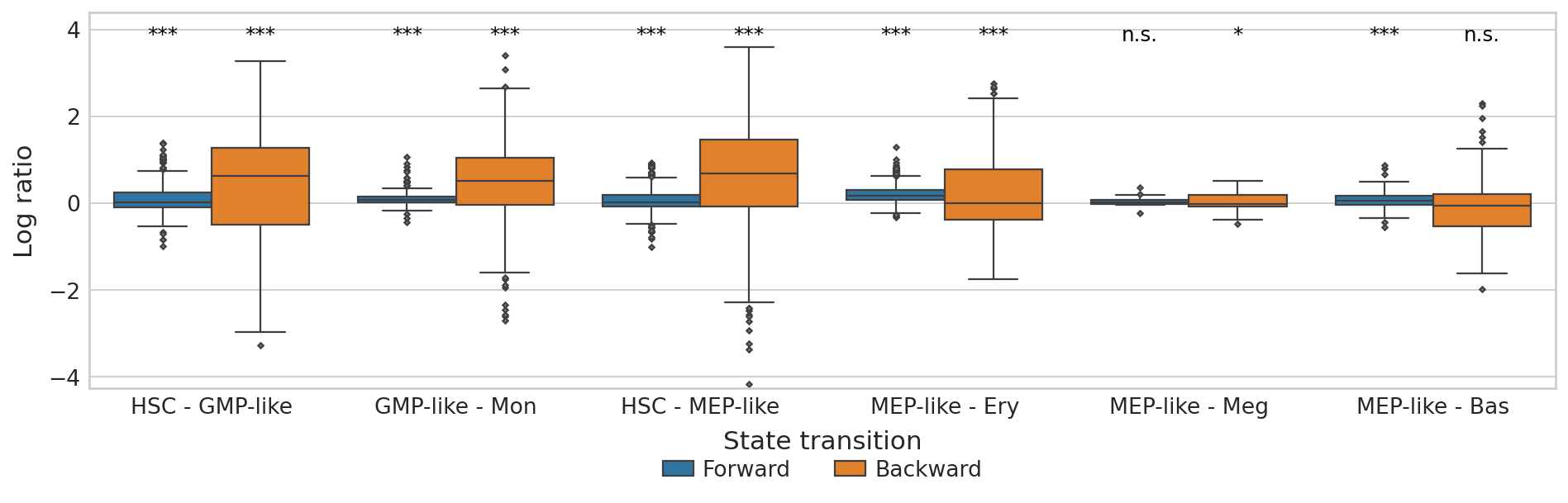
Plot CBC#
score_forward = []
for i in list(significances_forward.keys()):
score_forward.append(significances_forward[i])
score_backward = []
for i in list(significances_backward.keys()):
score_backward.append(significances_backward[i])
sig_m = pd.DataFrame(
{
"State transition": list(significances_forward.keys()) + list(significances_backward.keys()),
"Direction": ["Forward"] * 6 + ["Backward"] * 6,
"Sign": score_forward + score_backward,
}
)
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(1, 1, figsize=(12, 4), sharey=True)
flierprops = {"marker": "D", "markerfacecolor": "grey", "linestyle": "none", "markersize": 2}
sns.boxplot(x="State transition", y="Log ratio", hue="Direction", data=score_df, flierprops=flierprops, ax=ax)
# Calculate the maximum Y value from the data to set a uniform height for significance indicators
y_max = score_df["Log ratio"].max()
# Add significance indicators
cell_types = score_df["State transition"].unique()
categories = score_df["Direction"].unique()
for i, cell_type in enumerate(cell_types):
for j, category in enumerate(categories):
star = sig_m[(sig_m["State transition"].isin([cell_type])) & (sig_m["Direction"].isin([category]))][
"Sign"
].tolist()[0]
# Calculate the position of the asterisk
x = i + (j - 0.5) * 0.4 # Adjust the position of the asterisk
y = y_max + 0.05 # Uniform height for all asterisks
ax.text(x, y, star, ha="center", va="bottom", color="black")
plt.ylim(score_df["Log ratio"].min() - 0.1, score_df["Log ratio"].max() + 0.8)
plt.legend(loc="lower center", bbox_to_anchor=(0.5, -0.3), ncol=len(categories))
# Apply tight layout
plt.tight_layout()
if SAVE_FIGURES:
fig.savefig(
FIG_DIR / DATASET / "transition_benchmark_velocity.svg", format="svg", transparent=True, bbox_inches="tight"
)
plt.show()
Comparing RegVelo and scVelo#
Measuring forward CBC#
cluster_key = "cell_type"
rep = "X_pca"
score_df = []
for source, target in tqdm(STATE_TRANSITIONS):
cbc_rgv = vks["regvelo"].cbc(source=source, target=target, cluster_key=cluster_key, rep=rep)
cbc_scv = vks["scvelo"].cbc(source=source, target=target, cluster_key=cluster_key, rep=rep)
score_df.append(
pd.DataFrame(
{
"State transition": [f"{source} - {target}"] * len(cbc_rgv),
"Log ratio": np.log((cbc_rgv + 1) / (cbc_scv + 1)),
}
)
)
score_df_forward = pd.concat(score_df)
100%|██████████| 6/6 [00:00<00:00, 6.02it/s]
dfs = []
ttest_res = {}
significances = {}
for source, target in STATE_TRANSITIONS:
obs_mask = score_df_forward["State transition"].isin([f"{source} - {target}"])
a = score_df_forward.loc[obs_mask, "Log ratio"].values
b = np.zeros(len(a))
ttest_res[f"{source} - {target}"] = ttest_ind(a, b, equal_var=False, alternative="greater")
significances[f"{source} - {target}"] = get_significance(ttest_res[f"{source} - {target}"].pvalue)
significances_forward = significances.copy()
significances_forward
{'HSC - GMP-like': '***',
'GMP-like - Mon': '***',
'HSC - MEP-like': 'n.s.',
'MEP-like - Ery': '***',
'MEP-like - Meg': 'n.s.',
'MEP-like - Bas': '**'}
Measuring backward CBC#
score_df = []
for source, target in tqdm(STATE_TRANSITIONS_REVERSE):
cbc_rgv = -vks["regvelo"].cbc(source=source, target=target, cluster_key=cluster_key, rep=rep)
cbc_scv = -vks["scvelo"].cbc(source=source, target=target, cluster_key=cluster_key, rep=rep)
score_df.append(
pd.DataFrame(
{
"State transition": [f"{source} - {target}"] * len(cbc_rgv),
"Log ratio": np.log((cbc_rgv + 1) / (cbc_scv + 1)),
}
)
)
score_df_backward = pd.concat(score_df)
100%|██████████| 6/6 [00:00<00:00, 6.63it/s]
dfs = []
ttest_res = {}
significances = {}
for source, target in STATE_TRANSITIONS_REVERSE:
obs_mask = score_df_backward["State transition"].isin([f"{source} - {target}"])
a = score_df_backward.loc[obs_mask, "Log ratio"].values
b = np.zeros(len(a))
ttest_res[f"{source} - {target}"] = ttest_ind(a, b, equal_var=False, alternative="greater")
significances[f"{source} - {target}"] = get_significance(ttest_res[f"{source} - {target}"].pvalue)
significances_backward = significances.copy()
significances_backward
{'GMP-like - HSC': 'n.s.',
'Mon - GMP-like': 'n.s.',
'MEP-like - HSC': 'n.s.',
'Ery - MEP-like': '**',
'Meg - MEP-like': '***',
'Bas - MEP-like': '***'}
score_df_backward["State transition"] = score_df_backward["State transition"].apply(reverse_cluster)
score_df_forward.loc[:, "Direction"] = "Forward"
score_df_backward.loc[:, "Direction"] = "Backward"
score_df = pd.concat([score_df_forward, score_df_backward])
significances_backward = reverse_cluster_dict(significances_backward)
significances_backward
{'HSC - GMP-like': 'n.s.',
'GMP-like - Mon': 'n.s.',
'HSC - MEP-like': 'n.s.',
'MEP-like - Ery': '**',
'MEP-like - Meg': '***',
'MEP-like - Bas': '***'}
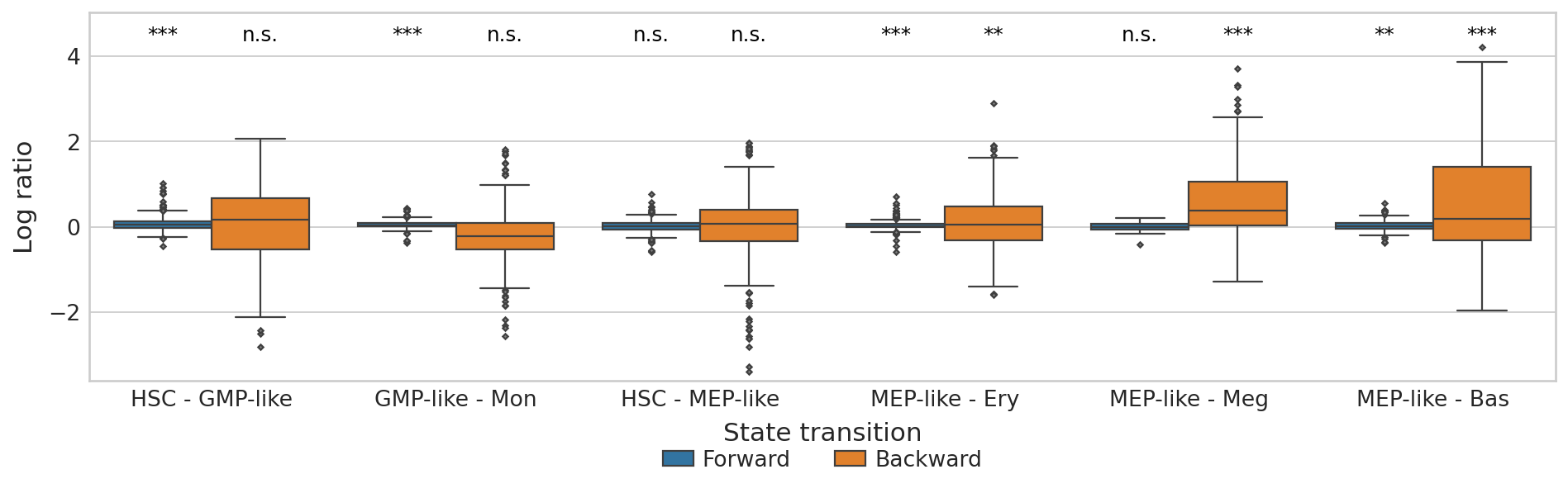
Plot CBC#
score_forward = []
for i in list(significances_forward.keys()):
score_forward.append(significances_forward[i])
score_backward = []
for i in list(significances_backward.keys()):
score_backward.append(significances_backward[i])
sig_m = pd.DataFrame(
{
"State transition": list(significances_forward.keys()) + list(significances_backward.keys()),
"Direction": ["Forward"] * 6 + ["Backward"] * 6,
"Sign": score_forward + score_backward,
}
)
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(1, 1, figsize=(12, 4), sharey=True)
flierprops = {"marker": "D", "markerfacecolor": "grey", "linestyle": "none", "markersize": 2}
sns.boxplot(x="State transition", y="Log ratio", hue="Direction", data=score_df, flierprops=flierprops, ax=ax)
# Calculate the maximum Y value from the data to set a uniform height for significance indicators
y_max = score_df["Log ratio"].max()
# Add significance indicators
cell_types = score_df["State transition"].unique()
categories = score_df["Direction"].unique()
for i, cell_type in enumerate(cell_types):
for j, category in enumerate(categories):
star = sig_m[(sig_m["State transition"].isin([cell_type])) & (sig_m["Direction"].isin([category]))][
"Sign"
].tolist()[0]
# Calculate the position of the asterisk
x = i + (j - 0.5) * 0.4 # Adjust the position of the asterisk
y = y_max + 0.05 # Uniform height for all asterisks
ax.text(x, y, star, ha="center", va="bottom", color="black")
plt.ylim(score_df["Log ratio"].min() - 0.2, score_df["Log ratio"].max() + 0.8)
plt.legend(loc="lower center", bbox_to_anchor=(0.5, -0.3), ncol=len(categories))
# Apply tight layout
plt.tight_layout()
if SAVE_FIGURES:
fig.savefig(FIG_DIR / DATASET / "rgv_scv.svg", format="svg", transparent=True, bbox_inches="tight")
plt.show()
Comparing RegVelo and veloVI#
Measuring forward CBC#
cluster_key = "cell_type"
rep = "X_pca"
score_df = []
for source, target in tqdm(STATE_TRANSITIONS):
cbc_rgv = vks["regvelo"].cbc(source=source, target=target, cluster_key=cluster_key, rep=rep)
cbc_vi = vks["velovi"].cbc(source=source, target=target, cluster_key=cluster_key, rep=rep)
score_df.append(
pd.DataFrame(
{
"State transition": [f"{source} - {target}"] * len(cbc_rgv),
"Log ratio": np.log((cbc_rgv + 1) / (cbc_vi + 1)),
}
)
)
score_df_forward = pd.concat(score_df)
100%|██████████| 6/6 [00:01<00:00, 5.98it/s]
dfs = []
ttest_res = {}
significances = {}
for source, target in STATE_TRANSITIONS:
obs_mask = score_df_forward["State transition"].isin([f"{source} - {target}"])
a = score_df_forward.loc[obs_mask, "Log ratio"].values
b = np.zeros(len(a))
ttest_res[f"{source} - {target}"] = ttest_ind(a, b, equal_var=False, alternative="greater")
significances[f"{source} - {target}"] = get_significance(ttest_res[f"{source} - {target}"].pvalue)
significances_forward = significances.copy()
significances_forward
{'HSC - GMP-like': 'n.s.',
'GMP-like - Mon': '***',
'HSC - MEP-like': 'n.s.',
'MEP-like - Ery': 'n.s.',
'MEP-like - Meg': 'n.s.',
'MEP-like - Bas': 'n.s.'}
Measuring backward CBC#
score_df = []
for source, target in tqdm(STATE_TRANSITIONS_REVERSE):
cbc_rgv = -vks["regvelo"].cbc(source=source, target=target, cluster_key=cluster_key, rep=rep)
cbc_vi = -vks["velovi"].cbc(source=source, target=target, cluster_key=cluster_key, rep=rep)
score_df.append(
pd.DataFrame(
{
"State transition": [f"{source} - {target}"] * len(cbc_rgv),
"Log ratio": np.log((cbc_rgv + 1) / (cbc_vi + 1)),
}
)
)
score_df_backward = pd.concat(score_df)
100%|██████████| 6/6 [00:00<00:00, 6.59it/s]
dfs = []
ttest_res = {}
significances = {}
for source, target in STATE_TRANSITIONS_REVERSE:
obs_mask = score_df_backward["State transition"].isin([f"{source} - {target}"])
a = score_df_backward.loc[obs_mask, "Log ratio"].values
b = np.zeros(len(a))
ttest_res[f"{source} - {target}"] = ttest_ind(a, b, equal_var=False, alternative="greater")
significances[f"{source} - {target}"] = get_significance(ttest_res[f"{source} - {target}"].pvalue)
significances_backward = significances.copy()
significances_backward
{'GMP-like - HSC': '***',
'Mon - GMP-like': '***',
'MEP-like - HSC': '***',
'Ery - MEP-like': '***',
'Meg - MEP-like': '**',
'Bas - MEP-like': '***'}
score_df_backward["State transition"] = score_df_backward["State transition"].apply(reverse_cluster)
score_df_forward.loc[:, "Direction"] = "Forward"
score_df_backward.loc[:, "Direction"] = "Backward"
score_df = pd.concat([score_df_forward, score_df_backward])
significances_backward = reverse_cluster_dict(significances_backward)
significances_backward
{'HSC - GMP-like': '***',
'GMP-like - Mon': '***',
'HSC - MEP-like': '***',
'MEP-like - Ery': '***',
'MEP-like - Meg': '**',
'MEP-like - Bas': '***'}
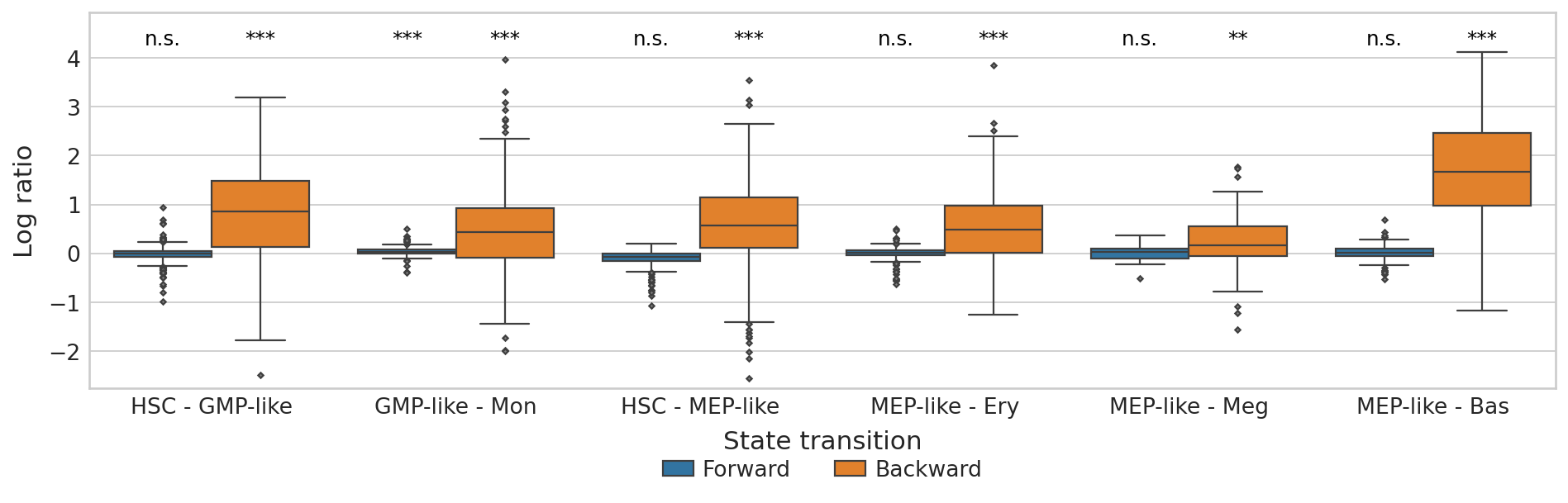
Plot CBC#
score_forward = []
for i in list(significances_forward.keys()):
score_forward.append(significances_forward[i])
score_backward = []
for i in list(significances_backward.keys()):
score_backward.append(significances_backward[i])
sig_m = pd.DataFrame(
{
"State transition": list(significances_forward.keys()) + list(significances_backward.keys()),
"Direction": ["Forward"] * 6 + ["Backward"] * 6,
"Sign": score_forward + score_backward,
}
)
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(1, 1, figsize=(12, 4), sharey=True)
flierprops = {"marker": "D", "markerfacecolor": "grey", "linestyle": "none", "markersize": 2}
sns.boxplot(x="State transition", y="Log ratio", hue="Direction", data=score_df, flierprops=flierprops, ax=ax)
# Calculate the maximum Y value from the data to set a uniform height for significance indicators
y_max = score_df["Log ratio"].max()
# Add significance indicators
cell_types = score_df["State transition"].unique()
categories = score_df["Direction"].unique()
for i, cell_type in enumerate(cell_types):
for j, category in enumerate(categories):
star = sig_m[(sig_m["State transition"].isin([cell_type])) & (sig_m["Direction"].isin([category]))][
"Sign"
].tolist()[0]
# Calculate the position of the asterisk
x = i + (j - 0.5) * 0.4 # Adjust the position of the asterisk
y = y_max + 0.05 # Uniform height for all asterisks
ax.text(x, y, star, ha="center", va="bottom", color="black")
plt.ylim(score_df["Log ratio"].min() - 0.2, score_df["Log ratio"].max() + 0.8)
plt.legend(loc="lower center", bbox_to_anchor=(0.5, -0.3), ncol=len(categories))
# Apply tight layout
plt.tight_layout()
plt.show()
if SAVE_FIGURES:
fig.savefig(FIG_DIR / DATASET / "rgv_velovi.svg", format="svg", transparent=True, bbox_inches="tight")
plt.show()
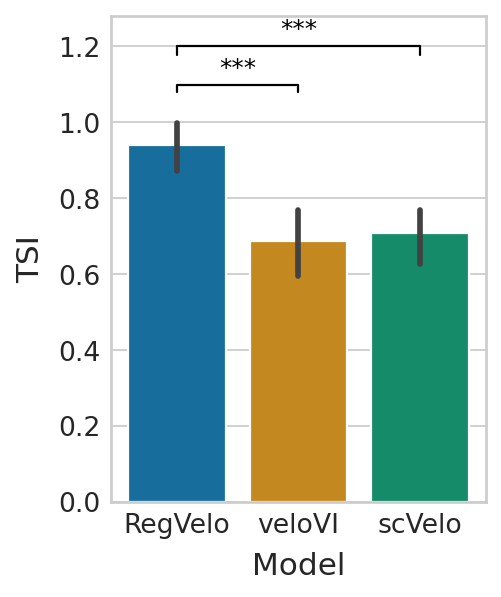
Terminal state identification#
ks = {}
for method in VELOCITY_METHODS:
adata = sc.read_h5ad(DATA_DIR / DATASET / "processed" / f"adata_run_{method}.h5ad")
## construct graph
ck = cr.kernels.ConnectivityKernel(adata).compute_transition_matrix()
vk = cr.kernels.VelocityKernel(adata)
vk.compute_transition_matrix()
ks[method] = 0.8 * vk + 0.2 * ck
thresholds = np.linspace(0.1, 1, 21)[:20]
estimators = {}
tsi = {}
for method in VELOCITY_METHODS[1:]:
estimators[method] = cr.estimators.GPCCA(vks[method])
tsi[method] = get_tsi_score(adata, thresholds, "cell_type", TERMINAL_STATES, estimators[method])
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Requested more macrostates `3` than available Schur vectors `2`. Recomputing the decomposition
WARNING: Requested more macrostates `4` than available Schur vectors `3`. Recomputing the decomposition
WARNING: Requested more macrostates `5` than available Schur vectors `4`. Recomputing the decomposition
WARNING: Requested more macrostates `6` than available Schur vectors `5`. Recomputing the decomposition
WARNING: Requested more macrostates `7` than available Schur vectors `6`. Recomputing the decomposition
WARNING: Requested more macrostates `8` than available Schur vectors `7`. Recomputing the decomposition
WARNING: Requested more macrostates `9` than available Schur vectors `8`. Recomputing the decomposition
WARNING: Requested more macrostates `10` than available Schur vectors `9`. Recomputing the decomposition
WARNING: Requested more macrostates `11` than available Schur vectors `10`. Recomputing the decomposition
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Requested more macrostates `3` than available Schur vectors `2`. Recomputing the decomposition
WARNING: Requested more macrostates `4` than available Schur vectors `3`. Recomputing the decomposition
WARNING: Requested more macrostates `5` than available Schur vectors `4`. Recomputing the decomposition
WARNING: Requested more macrostates `6` than available Schur vectors `5`. Recomputing the decomposition
WARNING: Requested more macrostates `7` than available Schur vectors `6`. Recomputing the decomposition
WARNING: Requested more macrostates `8` than available Schur vectors `7`. Recomputing the decomposition
WARNING: Requested more macrostates `9` than available Schur vectors `8`. Recomputing the decomposition
WARNING: Requested more macrostates `10` than available Schur vectors `9`. Recomputing the decomposition
WARNING: Requested more macrostates `11` than available Schur vectors `10`. Recomputing the decomposition
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'GMP-like': 29, 'HSC': 29}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'GMP-like': 29, 'HSC': 29}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'GMP-like': 29, 'HSC': 29}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'GMP-like': 29, 'HSC': 29}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'GMP-like': 29, 'HSC': 29}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'GMP-like': 29, 'HSC': 29}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'GMP-like': 29, 'HSC': 29}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'GMP-like': 29, 'HSC': 29}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'GMP-like': 29, 'HSC': 29}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'GMP-like': 29, 'HSC': 29}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'GMP-like': 29, 'HSC': 29}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'GMP-like': 29, 'HSC': 29}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'GMP-like': 29, 'HSC': 29}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'GMP-like': 29, 'HSC': 29}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'GMP-like': 29, 'HSC': 29}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'GMP-like': 29, 'HSC': 29}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'GMP-like': 29, 'HSC': 29}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'GMP-like': 29, 'HSC': 29}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'GMP-like': 29, 'HSC': 29}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'GMP-like': 29, 'HSC': 29}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Requested more macrostates `3` than available Schur vectors `2`. Recomputing the decomposition
WARNING: Requested more macrostates `4` than available Schur vectors `3`. Recomputing the decomposition
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: Requested more macrostates `6` than available Schur vectors `5`. Recomputing the decomposition
WARNING: Requested more macrostates `7` than available Schur vectors `6`. Recomputing the decomposition
WARNING: Requested more macrostates `8` than available Schur vectors `7`. Recomputing the decomposition
WARNING: Unable to compute macrostates with `n_states=8` because it will split complex conjugate eigenvalues. Using `n_states=9`
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: Requested more macrostates `10` than available Schur vectors `9`. Recomputing the decomposition
WARNING: Requested more macrostates `11` than available Schur vectors `10`. Recomputing the decomposition
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: Unable to compute macrostates with `n_states=8` because it will split complex conjugate eigenvalues. Using `n_states=9`
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: Unable to compute macrostates with `n_states=8` because it will split complex conjugate eigenvalues. Using `n_states=9`
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: Unable to compute macrostates with `n_states=8` because it will split complex conjugate eigenvalues. Using `n_states=9`
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: Unable to compute macrostates with `n_states=8` because it will split complex conjugate eigenvalues. Using `n_states=9`
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: Unable to compute macrostates with `n_states=8` because it will split complex conjugate eigenvalues. Using `n_states=9`
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: Unable to compute macrostates with `n_states=8` because it will split complex conjugate eigenvalues. Using `n_states=9`
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: Unable to compute macrostates with `n_states=8` because it will split complex conjugate eigenvalues. Using `n_states=9`
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: Unable to compute macrostates with `n_states=8` because it will split complex conjugate eigenvalues. Using `n_states=9`
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: Unable to compute macrostates with `n_states=8` because it will split complex conjugate eigenvalues. Using `n_states=9`
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: Unable to compute macrostates with `n_states=8` because it will split complex conjugate eigenvalues. Using `n_states=9`
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: Unable to compute macrostates with `n_states=8` because it will split complex conjugate eigenvalues. Using `n_states=9`
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: Unable to compute macrostates with `n_states=8` because it will split complex conjugate eigenvalues. Using `n_states=9`
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: Unable to compute macrostates with `n_states=8` because it will split complex conjugate eigenvalues. Using `n_states=9`
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: Unable to compute macrostates with `n_states=8` because it will split complex conjugate eigenvalues. Using `n_states=9`
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: Unable to compute macrostates with `n_states=8` because it will split complex conjugate eigenvalues. Using `n_states=9`
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: Unable to compute macrostates with `n_states=8` because it will split complex conjugate eigenvalues. Using `n_states=9`
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: Unable to compute macrostates with `n_states=8` because it will split complex conjugate eigenvalues. Using `n_states=9`
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: Unable to compute macrostates with `n_states=8` because it will split complex conjugate eigenvalues. Using `n_states=9`
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: Unable to compute macrostates with `n_states=8` because it will split complex conjugate eigenvalues. Using `n_states=9`
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
df = pd.DataFrame(
{
"TSI": tsi["regvelo"] + tsi["velovi"] + tsi["scvelo"],
"Model": ["RegVelo"] * 20 + ["veloVI"] * 20 + ["scVelo"] * 20,
}
)
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(3, 4))
sns.barplot(data=df, x="Model", y="TSI", palette="colorblind", ax=ax)
ttest_res = ttest_ind(tsi["regvelo"], tsi["velovi"], alternative="greater")
significance = get_significance(ttest_res.pvalue)
add_significance(
ax=ax,
left=0,
right=1,
significance=significance,
lw=1,
bracket_level=1.05,
c="k",
level=0,
)
ttest_res = ttest_ind(tsi["regvelo"], tsi["scvelo"], alternative="greater")
significance = get_significance(ttest_res.pvalue)
add_significance(ax=ax, left=0, right=2, significance=significance, lw=1, c="k", level=2, bracket_level=0.9)
y_min, y_max = ax.get_ylim()
ax.set_ylim([y_min, y_max + 0.02])
if SAVE_FIGURES:
fig.savefig(FIG_DIR / DATASET / "TSI_benchmark_update.svg", format="svg", transparent=True, bbox_inches="tight")
plt.show()
Show the stair plot#
tsi_rgv_curve = plot_tsi(adata, estimators["regvelo"], 0.8, TERMINAL_STATES, "cell_type")
tsi_scv_curve = plot_tsi(adata, estimators["scvelo"], 0.8, TERMINAL_STATES, "cell_type")
tsi_vi_curve = plot_tsi(adata, estimators["velovi"], 0.8, TERMINAL_STATES, "cell_type")
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=11` because it will split complex conjugate eigenvalues. Using `n_states=12`
WARNING: The following terminal states have different number of cells than requested (30): {'GMP-like': 29, 'HSC': 29}
WARNING: Found only one macrostate, making it the singular terminal state
WARNING: Unable to compute macrostates with `n_states=4` because it will split complex conjugate eigenvalues. Using `n_states=5`
WARNING: Unable to compute macrostates with `n_states=8` because it will split complex conjugate eigenvalues. Using `n_states=9`
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
WARNING: The following terminal states have different number of cells than requested (30): {'Mon_2': 27}
### plot the recovery figure
df = pd.DataFrame(
{
"number_macrostate": range(0, 12),
"RegVelo": [0] + tsi_rgv_curve,
"veloVI": [0] + tsi_vi_curve,
"scVelo": [0] + tsi_scv_curve,
}
)
df = pd.melt(df, ["number_macrostate"])
# Set figure size
with mplscience.style_context():
sns.set_style(style="whitegrid")
rcParams["figure.figsize"] = 4, 3
# Plot the grid plot
ax = sns.lineplot(
x="number_macrostate",
y="value",
hue="variable",
style="variable",
palette=METHOD_PALETTE_TSI,
drawstyle="steps-post",
data=df,
linewidth=3,
)
# Set labels and titles
ax.set(ylabel="Number of correct predictions")
plt.xlabel("Number of macrostates", fontsize=14)
plt.ylabel("Identified terminal states", fontsize=14)
# Customize tick parameters for better readability
ax.set_xticks([0, 2, 4, 6, 8, 10])
ax.set_xticklabels([0, 2, 4, 6, 8, 10])
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.legend(loc="upper center", bbox_to_anchor=(0.5, -0.18), shadow=True, ncol=4, fontsize=14)
if SAVE_FIGURES:
plt.savefig(FIG_DIR / DATASET / "state_identification.svg", format="svg", transparent=True, bbox_inches="tight")
plt.show()