Performance comparison of inference on toy GRN data#
Notebook compares metrics for velocity, latent time and GRN inference across different methods applied to toy GRN data.
Library imports#
import pandas as pd
from scipy.stats import ttest_ind
import matplotlib.pyplot as plt
import mplscience
import seaborn as sns
from rgv_tools import DATA_DIR, FIG_DIR
from rgv_tools.core import METHOD_PALETTE
from rgv_tools.plotting._significance import add_significance, get_significance
General settings#
SAVE_FIGURES = False
if SAVE_FIGURES:
(FIG_DIR / "toy_grn").mkdir(parents=True, exist_ok=True)
FIGURE_FORMATE = "svg"
Constants#
VELOCITY_METHODS = ["regvelo", "velovi", "scvelo"]
TIME_METHODS = ["regvelo", "velovi", "scvelo", "dpt"]
GRN_METHODS = ["regvelo", "correlation", "grnboost2", "celloracle"]
Data loading#
correlation_df = []
for method in VELOCITY_METHODS:
df = pd.read_parquet(DATA_DIR / "toy_grn" / "results" / f"{method}_correlation.parquet")
df.columns = f"{method}_" + df.columns
correlation_df.append(df)
del df
for method in TIME_METHODS:
if method in VELOCITY_METHODS:
continue
df = pd.read_parquet(DATA_DIR / "toy_grn" / "results" / f"{method}_correlation.parquet")
df.columns = f"{method}_" + df.columns
correlation_df.append(df)
for method in GRN_METHODS:
if method in VELOCITY_METHODS + TIME_METHODS:
continue
df = pd.read_parquet(DATA_DIR / "toy_grn" / "results" / f"{method}_correlation.parquet")
df.columns = f"{method}_" + df.columns
correlation_df.append(df)
correlation_df = pd.concat(correlation_df, axis=1)
correlation_df.head()
| regvelo_velocity | regvelo_time | regvelo_grn | velovi_velocity | velovi_time | scvelo_velocity | scvelo_time | dpt_time | correlation_grn | grnboost2_grn | celloracle_grn | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.555112 | 0.564590 | 0.530 | 0.523382 | -0.462360 | 0.461017 | 0.466543 | -0.081997 | 0.59 | 0.475 | 0.470 |
| 1 | 0.747088 | 0.861632 | 0.725 | 0.565394 | 0.810599 | 0.621778 | 0.779928 | -0.008727 | 0.56 | 0.530 | 0.555 |
| 2 | 0.747022 | 0.786513 | 0.550 | 0.719559 | 0.613784 | 0.710150 | 0.833685 | 0.012410 | 0.71 | 0.565 | 0.520 |
| 3 | 0.795365 | 0.604589 | 0.530 | 0.706493 | 0.125379 | 0.765667 | 0.678434 | 0.824321 | 0.62 | 0.620 | 0.500 |
| 4 | 0.696613 | 0.914910 | 0.830 | 0.680337 | 0.199152 | 0.646673 | 0.718848 | -0.102730 | 0.69 | 0.550 | 0.595 |
Analysis#
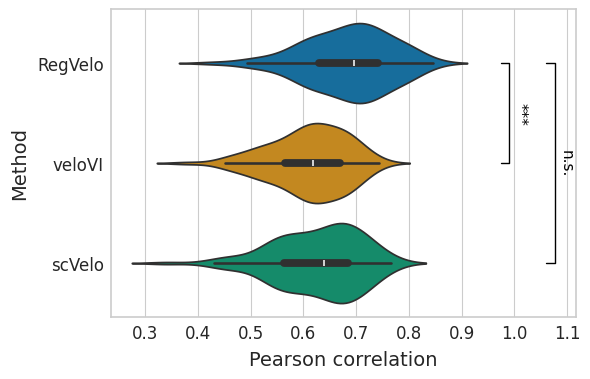
Velocity#
df = correlation_df.loc[:, correlation_df.columns.str.contains("velocity")]
df.columns = df.columns.str.removesuffix("_velocity")
df = pd.melt(df, var_name="method", value_name="correlation")
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(6, 4))
sns.violinplot(
data=df, x="correlation", y="method", hue="method", order=VELOCITY_METHODS, palette=METHOD_PALETTE, ax=ax
)
ttest_res = ttest_ind(
correlation_df["regvelo_velocity"],
correlation_df["velovi_velocity"],
equal_var=False,
alternative="greater",
)
significance = get_significance(pvalue=ttest_res.pvalue)
add_significance(
ax=ax,
left=0,
right=1,
significance=significance,
lw=1,
bracket_level=1.05,
c="k",
level=0,
orientation="vertical",
)
ttest_res = ttest_ind(
correlation_df["regvelo_velocity"],
correlation_df["scvelo_velocity"],
equal_var=False,
alternative="greater",
)
significance = get_significance(pvalue=ttest_res.pvalue)
add_significance(
ax=ax,
left=0,
right=2,
significance=significance,
lw=1,
bracket_level=1.05,
c="k",
level=0,
orientation="vertical",
)
ax.set(
xlabel="Pearson correlation",
ylabel="Method",
yticks=ax.get_yticks(),
yticklabels=["RegVelo", "veloVI", "scVelo"],
)
if SAVE_FIGURES:
fig.savefig(
FIG_DIR / "toy_GRN" / "velocity_benchmark.svg",
format="svg",
transparent=True,
bbox_inches="tight",
)
plt.show()
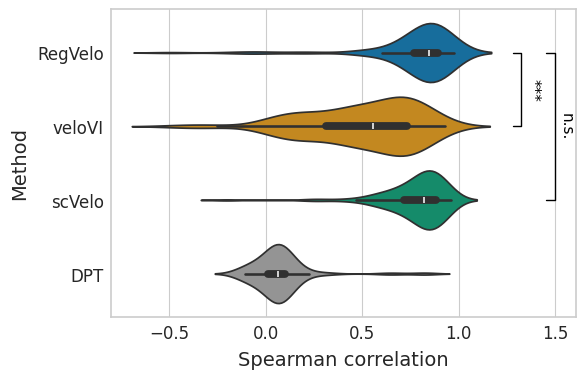
Latent time#
df = correlation_df.loc[:, correlation_df.columns.str.contains("time")]
df.columns = df.columns.str.removesuffix("_time")
df = pd.melt(df, var_name="method", value_name="correlation")
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(6, 4))
sns.violinplot(
data=df, x="correlation", y="method", hue="method", order=TIME_METHODS, palette=METHOD_PALETTE, ax=ax
)
ttest_res = ttest_ind(
correlation_df["regvelo_time"],
correlation_df["velovi_time"],
equal_var=False,
alternative="greater",
)
significance = get_significance(pvalue=ttest_res.pvalue)
add_significance(
ax=ax,
left=0,
right=1,
significance=significance,
lw=1,
bracket_level=1.05,
c="k",
level=0,
orientation="vertical",
)
ttest_res = ttest_ind(
correlation_df["velovi_time"],
correlation_df["scvelo_time"],
equal_var=False,
alternative="greater",
)
significance = get_significance(pvalue=ttest_res.pvalue)
add_significance(
ax=ax,
left=0,
right=2,
significance=significance,
lw=1,
bracket_level=1.05,
c="k",
level=0,
orientation="vertical",
)
ax.set(
xlabel="Spearman correlation",
ylabel="Method",
yticks=ax.get_yticks(),
yticklabels=["RegVelo", "veloVI", "scVelo", "DPT"],
)
if SAVE_FIGURES:
fig.savefig(
FIG_DIR / "toy_GRN" / "time_benchmark.svg",
format="svg",
transparent=True,
bbox_inches="tight",
)
plt.show()
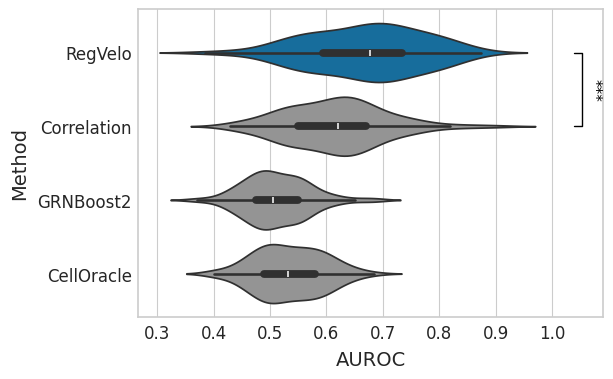
GRN#
df = correlation_df.loc[:, correlation_df.columns.str.contains("grn")]
df.columns = df.columns.str.removesuffix("_grn")
df = pd.melt(df, var_name="method").rename(columns={"value": "correlation"})
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(6, 4))
sns.violinplot(data=df, x="correlation", y="method", hue="method", order=GRN_METHODS, palette=METHOD_PALETTE, ax=ax)
ttest_res = ttest_ind(
correlation_df["regvelo_grn"],
correlation_df["correlation_grn"],
equal_var=False,
alternative="greater",
)
significance = get_significance(pvalue=ttest_res.pvalue)
add_significance(
ax=ax,
left=0,
right=1,
significance=significance,
lw=1,
bracket_level=1.05,
c="k",
level=0,
orientation="vertical",
)
ax.set(
xlabel="AUROC",
ylabel="Method",
yticks=ax.get_yticks(),
yticklabels=["RegVelo", "Correlation", "GRNBoost2", "CellOracle"],
)
if SAVE_FIGURES:
fig.savefig(
FIG_DIR / "toy_GRN" / "grn_benchmark.svg",
format="svg",
transparent=True,
bbox_inches="tight",
)
plt.show()