elf1 perturbation simulation#
Notebook for analyzing elf1 perturbation effects and regulatory circut
Library imports#
import numpy as np
import pandas as pd
import scipy
import matplotlib.pyplot as plt
import mplscience
import seaborn as sns
import cellrank as cr
import scanpy as sc
import scvelo as scv
from regvelo import REGVELOVI
from rgv_tools import DATA_DIR, FIG_DIR
from rgv_tools.benchmarking import set_output
from rgv_tools.perturbation import (
abundance_test,
get_list_name,
in_silico_block_simulation,
inferred_GRN,
RegulationScanning,
)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_csv from `anndata` is deprecated. Import anndata.io.read_csv instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_text from `anndata` is deprecated. Import anndata.io.read_text instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_excel from `anndata` is deprecated. Import anndata.io.read_excel instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_mtx from `anndata` is deprecated. Import anndata.io.read_mtx instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_loom from `anndata` is deprecated. Import anndata.io.read_loom instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_hdf from `anndata` is deprecated. Import anndata.io.read_hdf instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_umi_tools from `anndata` is deprecated. Import anndata.io.read_umi_tools instead.
warnings.warn(msg, FutureWarning)
General settings#
%matplotlib inline
plt.rcParams["svg.fonttype"] = "none"
sns.reset_defaults()
sns.reset_orig()
scv.settings.set_figure_params("scvelo", dpi_save=400, dpi=80, transparent=True, fontsize=14, color_map="viridis")
Constants#
DATASET = "zebrafish"
SAVE_DATA = True
if SAVE_DATA:
(DATA_DIR / DATASET / "results").mkdir(parents=True, exist_ok=True)
SAVE_FIGURES = False
if SAVE_FIGURES:
(FIG_DIR / DATASET).mkdir(parents=True, exist_ok=True)
TERMINAL_STATES = [
"mNC_head_mesenchymal",
"mNC_arch2",
"mNC_hox34",
"Pigment",
]
Data loading#
adata = sc.read_h5ad(DATA_DIR / DATASET / "processed" / "adata_run_regvelo.h5ad")
elf1 perturbation simulation#
Model loading#
model = DATA_DIR / DATASET / "processed" / "rgv_model"
vae = REGVELOVI.load(model, adata)
set_output(adata, vae)
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
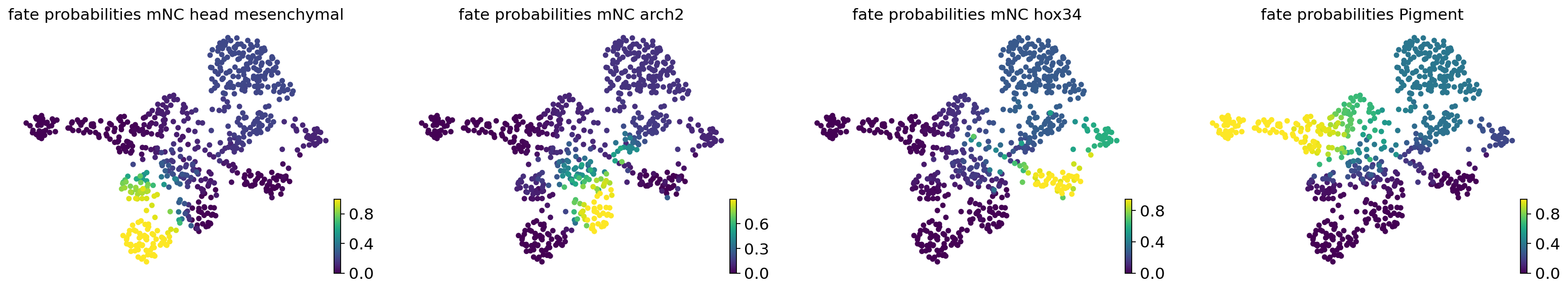
Calculate cell fate probabilities on original vector field#
vk = cr.kernels.VelocityKernel(adata)
vk.compute_transition_matrix()
ck = cr.kernels.ConnectivityKernel(adata).compute_transition_matrix()
estimator = cr.estimators.GPCCA(0.8 * vk + 0.2 * ck)
## evaluate the fate prob on original space
estimator.compute_macrostates(n_states=7, cluster_key="cell_type")
estimator.set_terminal_states(TERMINAL_STATES)
estimator.compute_fate_probabilities()
estimator.plot_fate_probabilities(same_plot=False)
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:12:33,406 - INFO - Using pre-computed Schur decomposition
WARNING: Unable to import petsc4py. For installation, please refer to: https://petsc4py.readthedocs.io/en/stable/install.html.
Defaulting to `'gmres'` solver.
Calculate cell fate probabilities on perturbed vector field#
adata_target_perturb, reg_vae_perturb = in_silico_block_simulation(model, adata, "elf1")
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
vk = cr.kernels.VelocityKernel(adata_target_perturb)
vk.compute_transition_matrix()
ck = cr.kernels.ConnectivityKernel(adata_target_perturb).compute_transition_matrix()
estimator_p = cr.estimators.GPCCA(0.8 * vk + 0.2 * ck)
## evaluate the fate prob on original space
estimator_p.compute_macrostates(n_states=7, cluster_key="cell_type")
estimator_p.set_terminal_states(TERMINAL_STATES)
estimator_p.compute_fate_probabilities()
## visualize coefficient
cond1_df = pd.DataFrame(
adata_target_perturb.obsm["lineages_fwd"], columns=adata_target_perturb.obsm["lineages_fwd"].names.tolist()
)
cond2_df = pd.DataFrame(adata.obsm["lineages_fwd"], columns=adata.obsm["lineages_fwd"].names.tolist())
## abundance test
data = abundance_test(cond2_df, cond1_df)
data = pd.DataFrame(
{
"Score": data.iloc[:, 0].tolist(),
"p-value": data.iloc[:, 1].tolist(),
"Terminal state": data.index.tolist(),
"TF": ["elf1"] * (data.shape[0]),
}
)
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:12:42,665 - INFO - Using pre-computed Schur decomposition
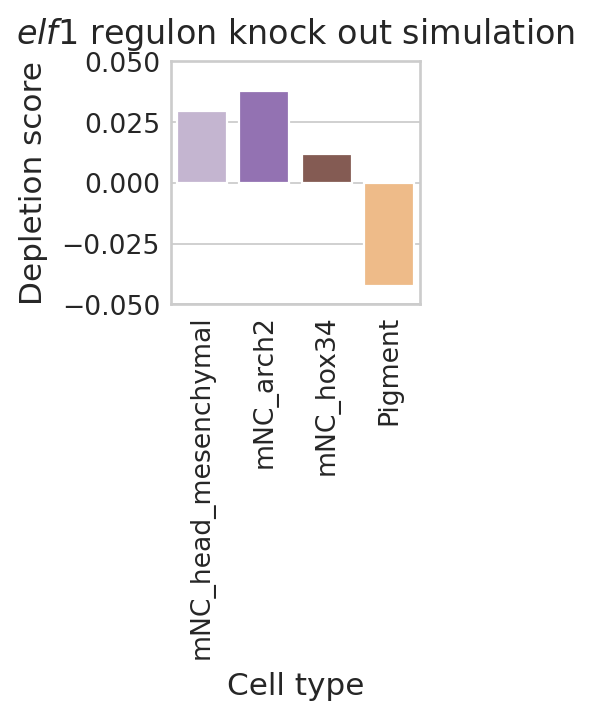
final_df = data.copy()
final_df["Score"] = 0.5 - final_df["Score"]
color_label = "cell_type"
df = pd.DataFrame(final_df["Score"])
df.columns = ["Depletion score"]
df["Cell type"] = final_df["Terminal state"]
order = df["Cell type"].tolist()
palette = dict(zip(adata.obs[color_label].cat.categories, adata.uns[f"{color_label}_colors"]))
subset_palette = {name: color for name, color in palette.items() if name in final_df.loc[:, "Terminal state"].tolist()}
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(2, 2))
sns.barplot(
data=df,
y="Depletion score",
x="Cell type",
palette=subset_palette,
order=order,
ax=ax,
)
ax.set(ylim=(-0.05, 0.05))
ax.tick_params(axis="x", rotation=90)
plt.title("$\\mathit{" + "elf1" + "}$ regulon knock out simulation")
if SAVE_FIGURES:
plt.savefig(
FIG_DIR / DATASET / "elf1_perturbation_simulation.svg", format="svg", transparent=True, bbox_inches="tight"
)
# Show the plot
plt.show()
GRN computation#
GRN = inferred_GRN(vae, adata, label="cell_type", group="all", data_frame=True)
Computing global GRN...
elf1 target screening#
targets = GRN.loc[:, "elf1"]
targets = np.array(targets.index.tolist())[np.array(targets) != 0]
print("inferring perturbation...")
perturb_screening = RegulationScanning(model, adata, 7, "cell_type", TERMINAL_STATES, "elf1", targets, 0)
coef = pd.DataFrame(np.array(perturb_screening["coefficient"]))
coef.index = perturb_screening["target"]
coef.columns = get_list_name(perturb_screening["coefficient"][0])
inferring perturbation...
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:12:55,891 - INFO - Using pre-computed Schur decomposition
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:13:09,564 - INFO - Using pre-computed Schur decomposition
Done ENSDARG00000024966
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:13:22,520 - INFO - Using pre-computed Schur decomposition
Done ENSDARG00000042329
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:13:35,339 - INFO - Using pre-computed Schur decomposition
Done alcama
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:13:48,245 - INFO - Using pre-computed Schur decomposition
Done aopep
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:14:01,784 - INFO - Using pre-computed Schur decomposition
Done apc
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:14:15,353 - INFO - Using pre-computed Schur decomposition
Done atp6v0ca
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:14:28,892 - INFO - Using pre-computed Schur decomposition
Done baz1b
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:14:43,256 - INFO - Using pre-computed Schur decomposition
Done calr3b
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:14:56,582 - INFO - Using pre-computed Schur decomposition
Done ccny
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:15:10,068 - INFO - Using pre-computed Schur decomposition
Done cdk1
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:15:23,049 - INFO - Using pre-computed Schur decomposition
Done cdkn1ca
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:15:35,974 - INFO - Using pre-computed Schur decomposition
Done celf2
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:15:48,925 - INFO - Using pre-computed Schur decomposition
Done cenpf
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:16:01,766 - INFO - Using pre-computed Schur decomposition
Done cpeb4b
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:16:15,175 - INFO - Using pre-computed Schur decomposition
Done cxxc5b
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:16:29,623 - INFO - Using pre-computed Schur decomposition
Done diaph2
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:16:42,563 - INFO - Using pre-computed Schur decomposition
Done dlg1
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:16:56,979 - INFO - Using pre-computed Schur decomposition
Done dnajb2
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:17:11,137 - INFO - Using pre-computed Schur decomposition
Done dnmt1
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:17:24,700 - INFO - Using pre-computed Schur decomposition
Done dusp5
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:17:37,726 - INFO - Using pre-computed Schur decomposition
Done ebf3a
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:17:51,116 - INFO - Using pre-computed Schur decomposition
Done emc2
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:18:04,107 - INFO - Using pre-computed Schur decomposition
Done ephb3a
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:18:17,957 - INFO - Using pre-computed Schur decomposition
Done erbb3b
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:18:31,203 - INFO - Using pre-computed Schur decomposition
Done esco2
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:18:44,101 - INFO - Using pre-computed Schur decomposition
Done ets1
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:18:57,224 - INFO - Using pre-computed Schur decomposition
Done eva1ba
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:19:10,181 - INFO - Using pre-computed Schur decomposition
Done fam49a
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:19:23,768 - INFO - Using pre-computed Schur decomposition
Done fhl3a
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:19:37,305 - INFO - Using pre-computed Schur decomposition
Done fhod1
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:19:51,016 - INFO - Using pre-computed Schur decomposition
Done fli1a
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:20:04,057 - INFO - Using pre-computed Schur decomposition
Done foxo1a
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:20:17,544 - INFO - Using pre-computed Schur decomposition
Done fzd3a
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:20:31,331 - INFO - Using pre-computed Schur decomposition
Done glb1l
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:20:44,414 - INFO - Using pre-computed Schur decomposition
Done glulb
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:20:57,600 - INFO - Using pre-computed Schur decomposition
Done gpd2
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:21:10,697 - INFO - Using pre-computed Schur decomposition
Done hat1
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:21:24,392 - INFO - Using pre-computed Schur decomposition
Done hexb
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:21:38,170 - INFO - Using pre-computed Schur decomposition
Done hivep1
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:21:51,188 - INFO - Using pre-computed Schur decomposition
Done hivep3b
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:22:04,155 - INFO - Using pre-computed Schur decomposition
Done hmgn2
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:22:17,260 - INFO - Using pre-computed Schur decomposition
Done hnrnpabb
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:22:30,930 - INFO - Using pre-computed Schur decomposition
Done hpcal4
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:22:43,842 - INFO - Using pre-computed Schur decomposition
Done hsp70.2
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:22:57,057 - INFO - Using pre-computed Schur decomposition
Done hspa5
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:23:10,085 - INFO - Using pre-computed Schur decomposition
Done hspb8
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:23:23,254 - INFO - Using pre-computed Schur decomposition
Done id2a
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:23:36,992 - INFO - Using pre-computed Schur decomposition
Done ildr2
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:23:50,141 - INFO - Using pre-computed Schur decomposition
Done inka1a
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:24:03,229 - INFO - Using pre-computed Schur decomposition
Done itga3a
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:24:17,712 - INFO - Using pre-computed Schur decomposition
Done itga8
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:24:33,000 - INFO - Using pre-computed Schur decomposition
Done ivns1abpa
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:24:46,854 - INFO - Using pre-computed Schur decomposition
Done klf6a
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:25:00,293 - INFO - Using pre-computed Schur decomposition
Done kntc1
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:25:13,596 - INFO - Using pre-computed Schur decomposition
Done mbnl2
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:25:26,944 - INFO - Using pre-computed Schur decomposition
Done metrn
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:25:40,431 - INFO - Using pre-computed Schur decomposition
Done mibp
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:25:53,708 - INFO - Using pre-computed Schur decomposition
Done myo10l1
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:26:06,849 - INFO - Using pre-computed Schur decomposition
Done nr6a1b
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:26:20,018 - INFO - Using pre-computed Schur decomposition
Done pcdh2g28
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:26:33,074 - INFO - Using pre-computed Schur decomposition
Done pdgfba
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:26:47,129 - INFO - Using pre-computed Schur decomposition
Done pdlim4
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:27:00,375 - INFO - Using pre-computed Schur decomposition
Done pleca
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:27:13,479 - INFO - Using pre-computed Schur decomposition
Done plpp3
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:27:26,802 - INFO - Using pre-computed Schur decomposition
Done pmp22a
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:27:40,425 - INFO - Using pre-computed Schur decomposition
Done ppt1
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:27:53,707 - INFO - Using pre-computed Schur decomposition
Done prdm1a
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:28:06,761 - INFO - Using pre-computed Schur decomposition
Done prkceb
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:28:20,094 - INFO - Using pre-computed Schur decomposition
Done prkcsh
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:28:33,164 - INFO - Using pre-computed Schur decomposition
Done pttg1ipb
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:28:47,076 - INFO - Using pre-computed Schur decomposition
Done rabl6b
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:29:00,421 - INFO - Using pre-computed Schur decomposition
Done ralgps2
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:29:14,788 - INFO - Using pre-computed Schur decomposition
Done rgl1
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:29:28,337 - INFO - Using pre-computed Schur decomposition
Done rhbdf1a
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:29:41,513 - INFO - Using pre-computed Schur decomposition
Done rhoca
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:29:55,226 - INFO - Using pre-computed Schur decomposition
Done rxraa
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:30:08,472 - INFO - Using pre-computed Schur decomposition
Done sash1b
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:30:21,732 - INFO - Using pre-computed Schur decomposition
Done sema3d
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:30:34,977 - INFO - Using pre-computed Schur decomposition
Done sema4ba
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:30:48,848 - INFO - Using pre-computed Schur decomposition
Done sept12
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:31:02,019 - INFO - Using pre-computed Schur decomposition
Done sept9a
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:31:15,226 - INFO - Using pre-computed Schur decomposition
Done serinc5
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:31:28,458 - INFO - Using pre-computed Schur decomposition
Done shroom4
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:31:41,715 - INFO - Using pre-computed Schur decomposition
Done si:ch211-199g17.2
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:31:56,148 - INFO - Using pre-computed Schur decomposition
Done si:ch211-222l21.1
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:32:09,345 - INFO - Using pre-computed Schur decomposition
Done si:ch73-335l21.1
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:32:24,218 - INFO - Using pre-computed Schur decomposition
Done si:dkey-17m8.1
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:32:37,533 - INFO - Using pre-computed Schur decomposition
Done sipa1l2
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:32:51,187 - INFO - Using pre-computed Schur decomposition
Done slbp
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:33:04,493 - INFO - Using pre-computed Schur decomposition
Done slc12a9
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:33:17,852 - INFO - Using pre-computed Schur decomposition
Done sox6
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:33:31,108 - INFO - Using pre-computed Schur decomposition
Done src
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:33:44,344 - INFO - Using pre-computed Schur decomposition
Done stat3
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:33:57,886 - INFO - Using pre-computed Schur decomposition
Done tcf12
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:34:11,078 - INFO - Using pre-computed Schur decomposition
Done tes
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:34:24,532 - INFO - Using pre-computed Schur decomposition
Done tle3b
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:34:37,771 - INFO - Using pre-computed Schur decomposition
Done tuba8l4
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:34:51,023 - INFO - Using pre-computed Schur decomposition
Done vash2
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:35:04,245 - INFO - Using pre-computed Schur decomposition
Done vav2
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:35:17,376 - INFO - Using pre-computed Schur decomposition
Done zgc:154093
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/zebrafish/processed/rgv_model/model.pt already
downloaded
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 17:35:30,736 - INFO - Using pre-computed Schur decomposition
Done zgc:165573
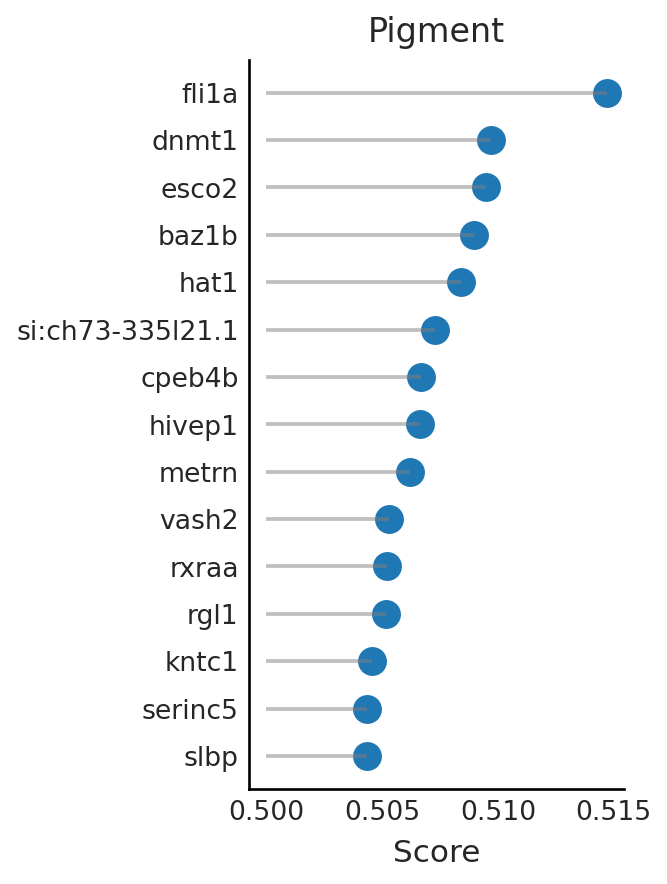
coef.sort_values("Pigment", ascending=False).iloc[:15,]
| mNC_head_mesenchymal | mNC_arch2 | mNC_hox34 | Pigment | |
|---|---|---|---|---|
| fli1a | 0.493243 | 0.474960 | 0.508461 | 0.514694 |
| dnmt1 | 0.497401 | 0.490594 | 0.496920 | 0.509698 |
| esco2 | 0.498142 | 0.487521 | 0.497025 | 0.509480 |
| baz1b | 0.497986 | 0.493608 | 0.496102 | 0.508951 |
| hat1 | 0.500184 | 0.489740 | 0.495598 | 0.508426 |
| si:ch73-335l21.1 | 0.497801 | 0.486049 | 0.499591 | 0.507288 |
| cpeb4b | 0.500248 | 0.491323 | 0.496722 | 0.506689 |
| hivep1 | 0.498229 | 0.489174 | 0.499295 | 0.506637 |
| metrn | 0.499143 | 0.487146 | 0.499159 | 0.506189 |
| vash2 | 0.498352 | 0.486737 | 0.501403 | 0.505310 |
| rxraa | 0.499764 | 0.503529 | 0.496063 | 0.505229 |
| rgl1 | 0.500238 | 0.492834 | 0.497344 | 0.505164 |
| kntc1 | 0.499017 | 0.497358 | 0.497949 | 0.504558 |
| serinc5 | 0.499229 | 0.489799 | 0.499768 | 0.504350 |
| slbp | 0.498465 | 0.493651 | 0.499795 | 0.504350 |
Pigment = coef.sort_values(by="Pigment", ascending=False)[:15]["Pigment"]
df = pd.DataFrame({"Gene": Pigment.index.tolist(), "Score": np.array(Pigment)})
# Sort DataFrame by -log10(p-value) for ordered plotting
df = df.sort_values(by="Score", ascending=False)
# Highlight specific genes
# Set up the plot
with mplscience.style_context():
sns.set_style(style="white")
fig, ax = plt.subplots(figsize=(3, 6))
sns.scatterplot(data=df, x="Score", y="Gene", palette="purple", s=200, legend=False)
for _, row in df.iterrows():
plt.hlines(row["Gene"], xmin=0.5, xmax=row["Score"], colors="grey", linestyles="-", alpha=0.5)
# Customize plot appearance
plt.xlabel("Score")
plt.ylabel("")
plt.title("Pigment")
plt.gca().spines["top"].set_visible(False)
plt.gca().spines["right"].set_visible(False)
plt.gca().spines["left"].set_color("black")
plt.gca().spines["bottom"].set_color("black")
# Show plot
if SAVE_FIGURES:
fig.savefig(FIG_DIR / DATASET / "elf1_driver.svg", format="svg", transparent=True, bbox_inches="tight")
plt.show()
Activity of positive regulated genes#
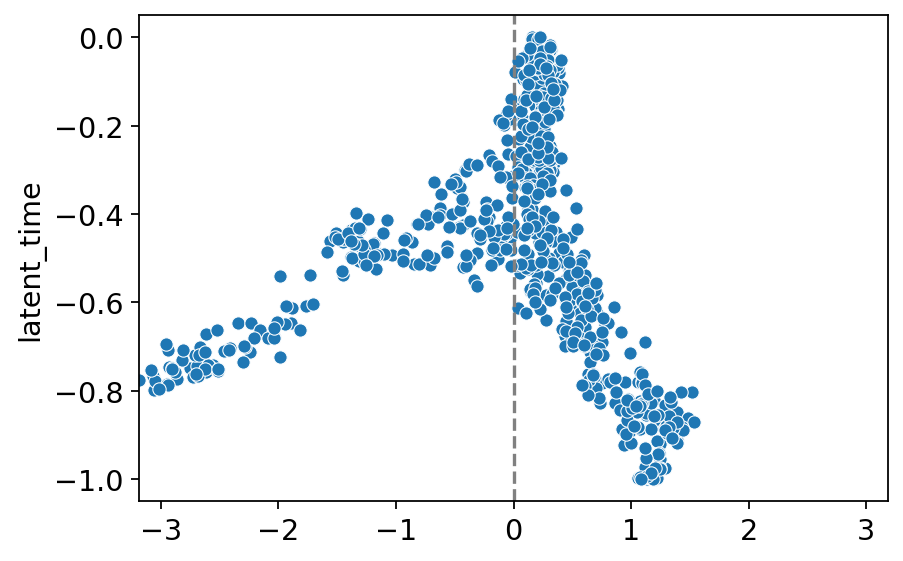
elf1_g = np.array(GRN.index.tolist())[np.array(GRN.loc[:, "elf1"]) != 0]
fli1a_g = np.array(GRN.index.tolist())[np.array(GRN.loc[:, "fli1a"]) != 0]
score = adata[:, elf1_g].layers["Ms"].mean(1) - adata[:, fli1a_g].layers["Ms"].mean(1)
score = scipy.stats.zscore(np.array(score))
sns.scatterplot(x=score, y=-adata.obs["latent_time"])
max_abs_x = max(abs(np.min(score)), abs(np.max(score)))
plt.xlim(-max_abs_x, max_abs_x)
# Display the plot
plt.axvline(0, color="grey", linestyle="--") # Optional: add a vertical line at x=0 for clarity
<matplotlib.lines.Line2D at 0x7f34e1175f00>
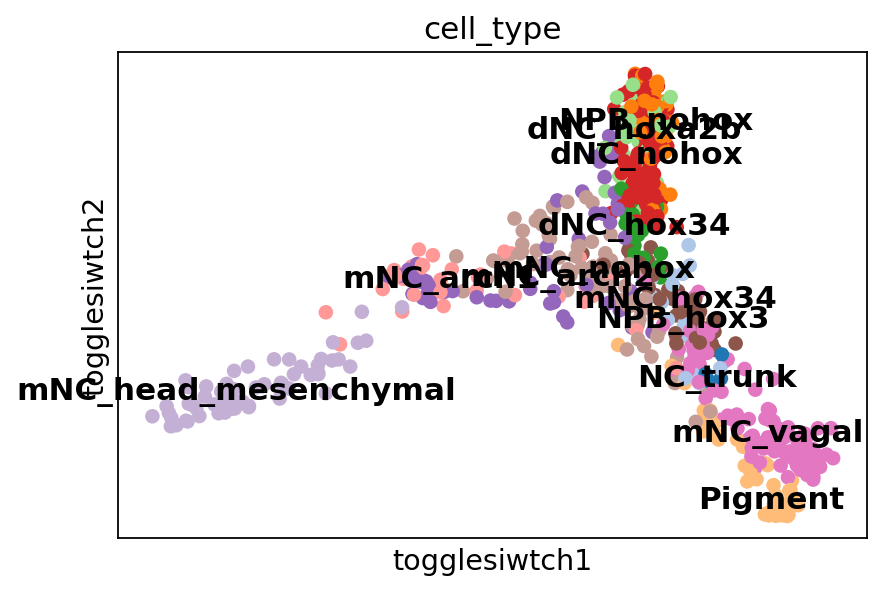
adata.obsm["X_togglesiwtch"] = np.column_stack((score, -adata.obs["latent_time"]))
sc.pl.embedding(adata, basis="togglesiwtch", color="cell_type", palette=sc.pl.palettes.vega_20, legend_loc="on data")
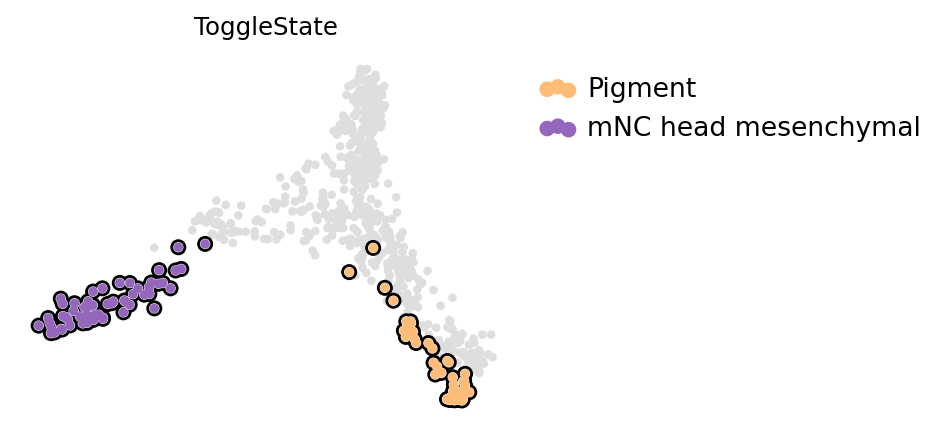
adata.obs["ToggleState"] = [i if i in ["mNC_head_mesenchymal", "Pigment"] else np.nan for i in adata.obs["macrostates"]]
adata.obs["ToggleState"] = adata.obs["ToggleState"].astype("category")
which = "ToggleState"
# adata.obs[which] = adata.obs["cell_type2"].copy()
state_names = adata.obs[which].cat.categories.tolist()
adata.obs[which] = adata.obs[which].astype(str).astype("category").cat.reorder_categories(["nan"] + state_names)
if which == "ToggleState":
adata.uns[f"{which}_colors"] = ["#dedede"] + list(subset_palette.values())
else:
adata.uns[f"{which}_colors"] = ["#dedede"] + list(subset_palette.values())
state_names = adata.obs[which].cat.categories.tolist()[1:]
with mplscience.style_context():
fig, ax = plt.subplots(figsize=(4, 3))
scv.pl.scatter(adata, basis="togglesiwtch", c=which, add_outline=state_names, ax=ax, size=60)
if SAVE_FIGURES:
fig.savefig(FIG_DIR / DATASET / "fli1a_elf1.svg", format="svg", transparent=True, bbox_inches="tight")
if SAVE_DATA:
adata.write_h5ad(DATA_DIR / DATASET / "results" / "elf1_screening.csv")