PU.1 and GATA1 perturbation#
Notebooks for PU.1 (SPI1) and GATA1 gene perturbation
Library imports#
import networkx as nx
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import mplscience
import seaborn as sns
from matplotlib.lines import Line2D
import cellrank as cr
import scanpy as sc
import scvelo as scv
from regvelo import REGVELOVI
from rgv_tools import DATA_DIR, FIG_DIR
from rgv_tools.benchmarking import set_output
from rgv_tools.perturbation import in_silico_block_simulation, inferred_GRN
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_csv from `anndata` is deprecated. Import anndata.io.read_csv instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_excel from `anndata` is deprecated. Import anndata.io.read_excel instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_hdf from `anndata` is deprecated. Import anndata.io.read_hdf instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_loom from `anndata` is deprecated. Import anndata.io.read_loom instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_mtx from `anndata` is deprecated. Import anndata.io.read_mtx instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_text from `anndata` is deprecated. Import anndata.io.read_text instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_umi_tools from `anndata` is deprecated. Import anndata.io.read_umi_tools instead.
warnings.warn(msg, FutureWarning)
General settings#
%matplotlib inline
plt.rcParams["svg.fonttype"] = "none"
scv.settings.set_figure_params("scvelo", dpi_save=400, dpi=80, transparent=True, fontsize=14, color_map="viridis")
Constants#
DATASET = "hematopoiesis"
SAVE_DATA = True
if SAVE_DATA:
(DATA_DIR / DATASET / "results").mkdir(parents=True, exist_ok=True)
SAVE_FIGURES = True
if SAVE_FIGURES:
(FIG_DIR / DATASET).mkdir(parents=True, exist_ok=True)
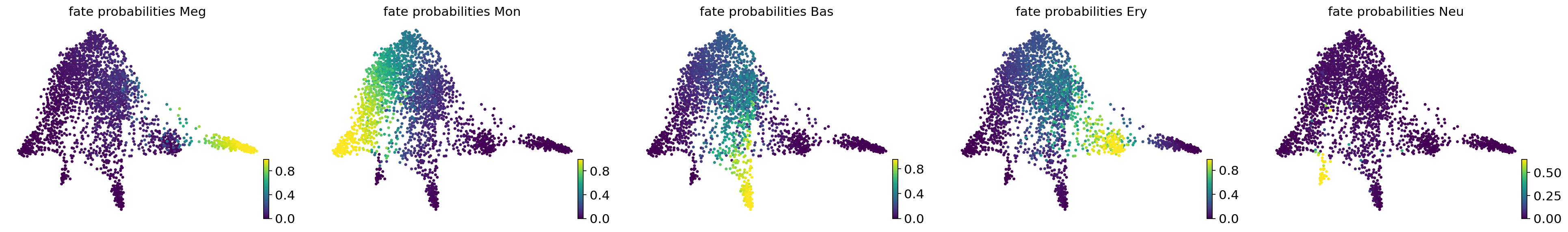
TERMINAL_STATES = ["Meg", "Mon", "Bas", "Ery", "Neu"]
Data loading#
adata = sc.read_h5ad(DATA_DIR / DATASET / "processed" / "adata_preprocessed.h5ad")
TF = adata.var_names[adata.var["TF"]]
PU.1 and GATA1 perturbation#
Model loading#
vae = REGVELOVI.load(DATA_DIR / DATASET / "processed" / "rgv_model", adata)
set_output(adata, vae, n_samples=30, batch_size=adata.n_obs)
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/hematopoiesis/processed/rgv_model/model.pt already
downloaded
vk = cr.kernels.VelocityKernel(adata)
vk.compute_transition_matrix()
ck = cr.kernels.ConnectivityKernel(adata).compute_transition_matrix()
estimator = cr.estimators.GPCCA(0.8 * vk + 0.2 * ck)
## evaluate the fate prob on original space
estimator.compute_macrostates(n_states=7, cluster_key="cell_type")
estimator.set_terminal_states(TERMINAL_STATES)
estimator.compute_fate_probabilities()
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 23:17:21,194 - INFO - Using pre-computed Schur decomposition
WARNING: Unable to import petsc4py. For installation, please refer to: https://petsc4py.readthedocs.io/en/stable/install.html.
Defaulting to `'gmres'` solver.
Knockout GATA1 simulation#
adata_perturb, vae_perturb = in_silico_block_simulation(
DATA_DIR / DATASET / "processed" / "rgv_model", adata, "GATA1", effects=0
)
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/hematopoiesis/processed/rgv_model/model.pt already
downloaded
vk = cr.kernels.VelocityKernel(adata_perturb)
vk.compute_transition_matrix()
ck = cr.kernels.ConnectivityKernel(adata_perturb).compute_transition_matrix()
estimator = cr.estimators.GPCCA(0.8 * vk + 0.2 * ck)
## evaluate the fate prob on original space
estimator.compute_macrostates(n_states=7, cluster_key="cell_type")
estimator.set_terminal_states(TERMINAL_STATES)
estimator.compute_fate_probabilities()
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 23:17:51,494 - INFO - Using pre-computed Schur decomposition
cond1_df = pd.DataFrame(
adata_perturb.obsm["lineages_fwd"], columns=adata_perturb.obsm["lineages_fwd"].names.tolist()
) # perturbed cell fate probabilities
cond2_df = pd.DataFrame(
adata.obsm["lineages_fwd"], columns=adata.obsm["lineages_fwd"].names.tolist()
) # original cell fate probabilities
## plot
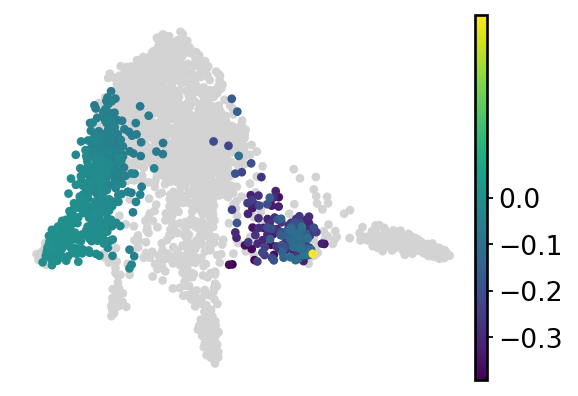
cell_fate = []
for i in range(cond1_df.shape[0]):
if cond2_df.iloc[i, 1] > 0.7 and np.abs(cond1_df.iloc[i, 1] - cond2_df.iloc[i, 1]) > 0:
cell_fate.append(cond1_df.iloc[i, 3] - cond2_df.iloc[i, 3])
elif cond2_df.iloc[i, 3] > 0.7 and np.abs(cond1_df.iloc[i, 3] - cond2_df.iloc[i, 3]) > 0:
cell_fate.append(cond1_df.iloc[i, 3] - cond2_df.iloc[i, 3])
else:
cell_fate.append(np.nan)
adata.obs["GATA1_perturb_effects"] = cell_fate
with mplscience.style_context():
fig, ax = plt.subplots(figsize=(4, 3))
sc.pl.embedding(
adata, basis="draw_graph_fa", color="GATA1_perturb_effects", frameon=False, title="", ax=ax, vcenter=0
)
if SAVE_FIGURES:
fig.savefig(FIG_DIR / DATASET / "GATA1_perturbation.svg", format="svg", transparent=True, bbox_inches="tight")
plt.show()
Knockout SPI1 simulation#
adata_perturb, vae_perturb = in_silico_block_simulation(
DATA_DIR / DATASET / "processed" / "rgv_model", adata, "SPI1", effects=0
)
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/hematopoiesis/processed/rgv_model/model.pt already
downloaded
vk = cr.kernels.VelocityKernel(adata_perturb)
vk.compute_transition_matrix()
ck = cr.kernels.ConnectivityKernel(adata_perturb).compute_transition_matrix()
estimator = cr.estimators.GPCCA(0.8 * vk + 0.2 * ck)
## evaluate the fate prob on original space
estimator.compute_macrostates(n_states=7, cluster_key="cell_type")
estimator.set_terminal_states(TERMINAL_STATES)
estimator.compute_fate_probabilities()
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
2024-11-25 23:19:28,030 - INFO - Using pre-computed Schur decomposition
cond1_df = pd.DataFrame(
adata_perturb.obsm["lineages_fwd"], columns=adata_perturb.obsm["lineages_fwd"].names.tolist()
) # perturbed cell fate probabilities
cond2_df = pd.DataFrame(
adata.obsm["lineages_fwd"], columns=adata.obsm["lineages_fwd"].names.tolist()
) # original cell fate probabilities
## plot
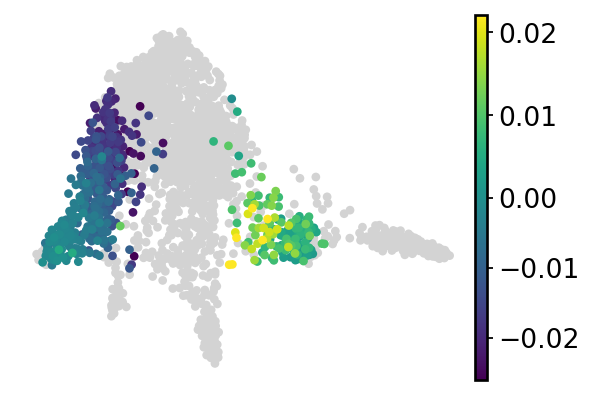
cell_fate = []
for i in range(cond1_df.shape[0]):
if cond2_df.iloc[i, 1] > 0.7 and np.abs(cond1_df.iloc[i, 1] - cond2_df.iloc[i, 1]) > 0:
cell_fate.append(cond1_df.iloc[i, 1] - cond2_df.iloc[i, 1])
elif cond2_df.iloc[i, 3] > 0.7 and np.abs(cond1_df.iloc[i, 3] - cond2_df.iloc[i, 3]) > 0:
cell_fate.append(cond1_df.iloc[i, 3] - cond2_df.iloc[i, 3])
else:
cell_fate.append(np.nan)
adata.obs["SPI1_perturb_effects"] = cell_fate
with mplscience.style_context():
fig, ax = plt.subplots(figsize=(4, 3))
sc.pl.embedding(
adata,
basis="draw_graph_fa",
color="SPI1_perturb_effects",
frameon=False,
vmin="p1",
vmax="p99",
title="",
ax=ax,
vcenter=0,
)
if SAVE_FIGURES:
fig.savefig(FIG_DIR / DATASET / "SPI1_perturbation.svg", format="svg", transparent=True, bbox_inches="tight")
plt.show()
Gene regulation motif between PU.1 and GATA1#
model = DATA_DIR / DATASET / "processed" / "perturb_repeat_runs" / "rgv_model_0"
vae = REGVELOVI.load(model, adata)
reg1 = inferred_GRN(vae, adata, label="cell_type", group=["Ery", "Meg"])
model = DATA_DIR / DATASET / "processed" / "perturb_repeat_runs" / "rgv_model_1"
vae = REGVELOVI.load(model, adata)
reg2 = inferred_GRN(vae, adata, label="cell_type", group=["Ery", "Meg"])
model = DATA_DIR / DATASET / "processed" / "perturb_repeat_runs" / "rgv_model_2"
vae = REGVELOVI.load(model, adata)
reg3 = inferred_GRN(vae, adata, label="cell_type", group=["Ery", "Meg"])
model = DATA_DIR / DATASET / "processed" / "perturb_repeat_runs" / "rgv_model_3"
vae = REGVELOVI.load(model, adata)
reg4 = inferred_GRN(vae, adata, label="cell_type", group=["Ery", "Meg"])
model = DATA_DIR / DATASET / "processed" / "perturb_repeat_runs" / "rgv_model_4"
vae = REGVELOVI.load(model, adata)
reg5 = inferred_GRN(vae, adata, label="cell_type", group=["Ery", "Meg"])
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/hematopoiesis/processed/perturb_repeat_runs/rgv_model_0
/model.pt already downloaded
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/hematopoiesis/processed/perturb_repeat_runs/rgv_model_1
/model.pt already downloaded
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/hematopoiesis/processed/perturb_repeat_runs/rgv_model_2
/model.pt already downloaded
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/hematopoiesis/processed/perturb_repeat_runs/rgv_model_3
/model.pt already downloaded
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:168: PossibleUserWarning: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
rank_zero_warn(
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:168: PossibleUserWarning: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
rank_zero_warn(
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:168: PossibleUserWarning: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
rank_zero_warn(
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:168: PossibleUserWarning: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
rank_zero_warn(
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/hematopoiesis/processed/perturb_repeat_runs/rgv_model_4
/model.pt already downloaded
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:168: PossibleUserWarning: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
rank_zero_warn(
regMotif = np.stack((reg1, reg2, reg3, reg4, reg5), axis=0)
regMotif = np.mean(regMotif, axis=0)
targets = regMotif[:, [i == "GATA1" for i in adata.var.index]].reshape(-1)
targets = pd.DataFrame(targets, index=adata.var.index)
targets.loc[:, "weight"] = targets.iloc[:, 0]
targets.sort_values("weight", ascending=False).iloc[:20, :]
| 0 | weight | |
|---|---|---|
| HBD | 22.301508 | 22.301508 |
| KEL | 11.961169 | 11.961169 |
| HBB | 11.001174 | 11.001174 |
| P2RX1 | 9.592329 | 9.592329 |
| RUFY1 | 8.332366 | 8.332366 |
| BLVRB | 6.952588 | 6.952588 |
| TAL1 | 6.601087 | 6.601087 |
| SLC37A1 | 5.963588 | 5.963588 |
| DAAM1 | 5.943871 | 5.943871 |
| STXBP5 | 5.741522 | 5.741522 |
| SPTA1 | 4.571942 | 4.571942 |
| TTC27 | 4.360682 | 4.360682 |
| GP6 | 4.252732 | 4.252732 |
| BTK | 4.163760 | 4.163760 |
| MTSS1 | 3.988803 | 3.988803 |
| CD36 | 3.865287 | 3.865287 |
| MYLK | 3.453176 | 3.453176 |
| ZFPM1 | 3.419088 | 3.419088 |
| PTGS1 | 2.836505 | 2.836505 |
| GFI1B | 2.806460 | 2.806460 |
model = DATA_DIR / DATASET / "processed" / "perturb_repeat_runs" / "rgv_model_0"
vae = REGVELOVI.load(model, adata)
reg1 = inferred_GRN(vae, adata, label="cell_type", group=["Neu", "Mon"])
model = DATA_DIR / DATASET / "processed" / "perturb_repeat_runs" / "rgv_model_1"
vae = REGVELOVI.load(model, adata)
reg2 = inferred_GRN(vae, adata, label="cell_type", group=["Neu", "Mon"])
model = DATA_DIR / DATASET / "processed" / "perturb_repeat_runs" / "rgv_model_2"
vae = REGVELOVI.load(model, adata)
reg3 = inferred_GRN(vae, adata, label="cell_type", group=["Neu", "Mon"])
model = DATA_DIR / DATASET / "processed" / "perturb_repeat_runs" / "rgv_model_3"
vae = REGVELOVI.load(model, adata)
reg4 = inferred_GRN(vae, adata, label="cell_type", group=["Neu", "Mon"])
model = DATA_DIR / DATASET / "processed" / "perturb_repeat_runs" / "rgv_model_4"
vae = REGVELOVI.load(model, adata)
reg5 = inferred_GRN(vae, adata, label="cell_type", group=["Neu", "Mon"])
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/hematopoiesis/processed/perturb_repeat_runs/rgv_model_0
/model.pt already downloaded
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/hematopoiesis/processed/perturb_repeat_runs/rgv_model_1
/model.pt already downloaded
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/hematopoiesis/processed/perturb_repeat_runs/rgv_model_2
/model.pt already downloaded
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:168: PossibleUserWarning: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
rank_zero_warn(
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:168: PossibleUserWarning: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
rank_zero_warn(
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:168: PossibleUserWarning: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
rank_zero_warn(
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:168: PossibleUserWarning: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
rank_zero_warn(
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/hematopoiesis/processed/perturb_repeat_runs/rgv_model_3
/model.pt already downloaded
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/hematopoiesis/processed/perturb_repeat_runs/rgv_model_4
/model.pt already downloaded
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:168: PossibleUserWarning: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
rank_zero_warn(
regMotif = np.stack((reg1, reg2, reg3, reg4, reg5), axis=0)
regMotif = np.mean(regMotif, axis=0)
targets = regMotif[:, [i == "SPI1" for i in adata.var.index]].reshape(-1)
targets = pd.DataFrame(targets, index=adata.var.index)
targets.loc[:, "weight"] = targets.iloc[:, 0]
targets.sort_values("weight", ascending=False).iloc[:20, :]
| 0 | weight | |
|---|---|---|
| SND1 | 13.434076 | 13.434076 |
| STT3B | 11.143422 | 11.143422 |
| HLA-DPB1 | 9.534171 | 9.534171 |
| ARHGAP30 | 8.252996 | 8.252996 |
| P2RY8 | 5.347001 | 5.347001 |
| TAF1 | 4.413145 | 4.413145 |
| CCDC88B | 3.921804 | 3.921804 |
| CD63 | 3.603989 | 3.603989 |
| NFIB | 3.186572 | 3.186572 |
| TCF4 | 2.952924 | 2.952924 |
| RREB1 | 2.627958 | 2.627958 |
| VIM | 2.054796 | 2.054796 |
| PPARA | 2.041915 | 2.041915 |
| MPO | 1.920108 | 1.920108 |
| FOXP1 | 1.686433 | 1.686433 |
| ZNF263 | 1.415900 | 1.415900 |
| ETV6 | 1.150037 | 1.150037 |
| ANXA1 | 1.076040 | 1.076040 |
| GATA2 | 0.927404 | 0.927404 |
| FTL | 0.621331 | 0.621331 |
Visualize global GRN#
model = DATA_DIR / DATASET / "processed" / "perturb_repeat_runs" / "rgv_model_0"
vae = REGVELOVI.load(model, adata)
reg1 = inferred_GRN(vae, adata, label="cell_type", group="all")
model = DATA_DIR / DATASET / "processed" / "perturb_repeat_runs" / "rgv_model_1"
vae = REGVELOVI.load(model, adata)
reg2 = inferred_GRN(vae, adata, label="cell_type", group="all")
model = DATA_DIR / DATASET / "processed" / "perturb_repeat_runs" / "rgv_model_2"
vae = REGVELOVI.load(model, adata)
reg3 = inferred_GRN(vae, adata, label="cell_type", group="all")
model = DATA_DIR / DATASET / "processed" / "perturb_repeat_runs" / "rgv_model_3"
vae = REGVELOVI.load(model, adata)
reg4 = inferred_GRN(vae, adata, label="cell_type", group="all")
model = DATA_DIR / DATASET / "processed" / "perturb_repeat_runs" / "rgv_model_4"
vae = REGVELOVI.load(model, adata)
reg5 = inferred_GRN(vae, adata, label="cell_type", group="all")
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/hematopoiesis/processed/perturb_repeat_runs/rgv_model_0
/model.pt already downloaded
Computing global GRN...
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/hematopoiesis/processed/perturb_repeat_runs/rgv_model_1
/model.pt already downloaded
Computing global GRN...
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/hematopoiesis/processed/perturb_repeat_runs/rgv_model_2
/model.pt already downloaded
Computing global GRN...
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/hematopoiesis/processed/perturb_repeat_runs/rgv_model_3
/model.pt already downloaded
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:168: PossibleUserWarning: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
rank_zero_warn(
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:168: PossibleUserWarning: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
rank_zero_warn(
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:168: PossibleUserWarning: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
rank_zero_warn(
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:168: PossibleUserWarning: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
rank_zero_warn(
Computing global GRN...
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/hematopoiesis/processed/perturb_repeat_runs/rgv_model_4
/model.pt already downloaded
Computing global GRN...
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:168: PossibleUserWarning: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
rank_zero_warn(
regMotif = np.stack((reg1, reg2, reg3, reg4, reg5), axis=0)
regMotif = np.mean(regMotif, axis=0)
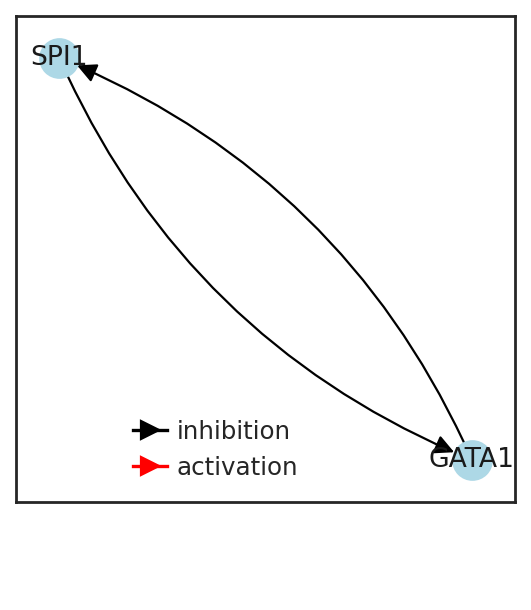
Plot toggle switch#
motif = [
["SPI1", "GATA1", regMotif[[i == "GATA1" for i in adata.var.index], [i == "SPI1" for i in adata.var.index]][0]],
["GATA1", "SPI1", regMotif[[i == "SPI1" for i in adata.var.index], [i == "GATA1" for i in adata.var.index]][0]],
]
motif = pd.DataFrame(motif)
motif.columns = ["from", "to", "weight"]
motif["weight"] = np.sign(motif["weight"])
legend_elements = [
Line2D([0], [0], marker=">", color="black", label="inhibition", markerfacecolor="black", markersize=8),
Line2D([0], [0], marker=">", color="red", label="activation", markerfacecolor="red", markersize=8),
]
with mplscience.style_context():
sns.set(style="white")
fig, ax = plt.subplots(figsize=(4, 4))
cont = motif
contLines = []
genes = set()
G = nx.MultiDiGraph()
pEdges = []
nEdges = []
for line in range(cont.shape[0]):
tmp = cont.iloc[line, :]
genes.add(tmp[0])
genes.add(tmp[1])
contLines.append(tmp.tolist())
genes = list(genes)
selfActGenes = set()
selfInhGenes = set()
G.add_nodes_from(genes)
for edge in contLines:
row = genes.index(edge[0])
col = genes.index(edge[1])
if edge[2] == 1:
pEdges.append((edge[0], edge[1]))
if row == col:
selfActGenes.add(edge[0])
elif edge[2] == -1:
nEdges.append((edge[0], edge[1]))
if row == col:
selfInhGenes.add(edge[0])
else:
print("Unsupported regulatory relationship.")
selfActGenes = list(selfActGenes)
selfInhGenes = list(selfInhGenes)
# show grn by network visualization
G.add_edges_from(pEdges)
G.add_edges_from(nEdges)
pos = nx.spring_layout(G)
nx.draw_networkx_nodes(G, pos, node_color="lightblue") # initial colors for all nodes
nx.draw_networkx_nodes(G, pos, nodelist=selfActGenes, node_color="red") # red colors indicating activation
nx.draw_networkx_nodes(G, pos, nodelist=selfInhGenes, node_color="black") # black colors indicating inhibition
nx.draw_networkx_labels(G, pos)
nx.draw_networkx_edges(G, pos, edgelist=pEdges, edge_color="red", connectionstyle="arc3,rad=0.2", arrowsize=18)
nx.draw_networkx_edges(
G, pos, edgelist=nEdges, edge_color="black", arrows=True, connectionstyle="arc3,rad=0.2", arrowsize=18
)
plt.legend(handles=legend_elements, bbox_to_anchor=(0.4, 0))
if SAVE_FIGURES:
fig.savefig(FIG_DIR / DATASET / "SPI1-GATA1-network.svg", format="svg", transparent=True, bbox_inches="tight")
plt.show()