Analyzing perturbation simulation results on pancreatic endocrine dataset#
Notebook for analyzing and visulizing RegVelo’s perturbation results
Library imports#
import numpy as np
import pandas as pd
from sklearn.metrics import auc, roc_curve
import matplotlib.pyplot as plt
import mplscience
import seaborn as sns
import scanpy as sc
import scvelo as scv
from rgv_tools import DATA_DIR, FIG_DIR
from rgv_tools.perturbation import aggregate_model_predictions
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_csv from `anndata` is deprecated. Import anndata.io.read_csv instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_excel from `anndata` is deprecated. Import anndata.io.read_excel instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_hdf from `anndata` is deprecated. Import anndata.io.read_hdf instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_loom from `anndata` is deprecated. Import anndata.io.read_loom instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_mtx from `anndata` is deprecated. Import anndata.io.read_mtx instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_text from `anndata` is deprecated. Import anndata.io.read_text instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_umi_tools from `anndata` is deprecated. Import anndata.io.read_umi_tools instead.
warnings.warn(msg, FutureWarning)
General settings#
%matplotlib inline
plt.rcParams["svg.fonttype"] = "none"
sns.reset_defaults()
sns.reset_orig()
scv.settings.set_figure_params("scvelo", dpi_save=400, dpi=80, transparent=True, fontsize=14, color_map="viridis")
Constants#
DATASET = "pancreatic_endocrinogenesis"
SAVE_DATA = True
if SAVE_DATA:
(DATA_DIR / DATASET / "results").mkdir(parents=True, exist_ok=True)
SAVE_FIGURES = True
if SAVE_FIGURES:
(FIG_DIR / DATASET).mkdir(parents=True, exist_ok=True)
TERMINAL_STATES = ["Alpha", "Beta", "Delta", "Epsilon"]
Function defination#
def min_max_scaling(data):
"""Apply min-max scaling to a numpy array or pandas Series.
Parameters
----------
data (np.ndarray or pd.Series): The input data to be scaled.
Returns
-------
np.ndarray or pd.Series: Scaled data with values between 0 and 1.
"""
min_val = np.min(data)
max_val = np.max(data)
scaled_data = (data - min_val) / (max_val - min_val)
return scaled_data
Data loading#
adata = sc.read_h5ad(DATA_DIR / DATASET / "processed" / "adata_preprocessed.h5ad")
regvelo_prediction = aggregate_model_predictions(DATA_DIR / DATASET / "results")
palette = dict(zip(adata.obs["clusters"].cat.categories, adata.uns["clusters_colors"]))
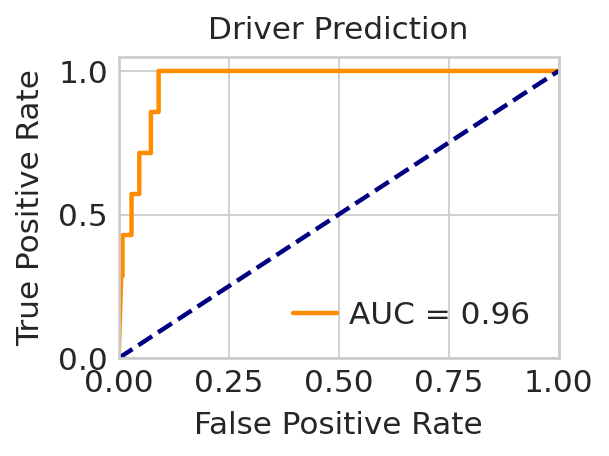
Drivers prediction#
driver_list = {
"Alpha": ["Pou6f2", "Irx1", "Irx2", "Smarca1"],
"Delta": ["Hhex"],
"Beta": ["Pdx1", "Glis3", "Mafa", "Mafb"],
"Epsilon": ["Arg4", "Nuak2", "Tead1"],
}
score = (
min_max_scaling(-regvelo_prediction[0].loc[:, "Alpha"]).tolist()
+ min_max_scaling(-regvelo_prediction[0].loc[:, "Beta"]).tolist()
+ min_max_scaling(-regvelo_prediction[0].loc[:, "Delta"]).tolist()
+ min_max_scaling(-regvelo_prediction[0].loc[:, "Epsilon"]).tolist()
)
label = (
[1 if i in driver_list["Alpha"] else 0 for i in regvelo_prediction[0].index.tolist()]
+ [1 if i in driver_list["Beta"] else 0 for i in regvelo_prediction[0].index.tolist()]
+ [1 if i in driver_list["Delta"] else 0 for i in regvelo_prediction[0].index.tolist()]
+ [1 if i in driver_list["Epsilon"] else 0 for i in regvelo_prediction[0].index.tolist()]
)
with mplscience.style_context(): # Entering the custom style context
# Calculate ROC curve and AUC
fpr, tpr, _ = roc_curve(label, score)
roc_auc = auc(fpr, tpr)
sns.set_style("whitegrid")
# Create a figure and axis object with a specified size
fig, ax = plt.subplots(figsize=(4, 3))
# Plot ROC curve
ax.plot(fpr, tpr, color="darkorange", lw=2, label=f"AUC = {roc_auc:.2f}")
# Plot diagonal line (random guess line)
ax.plot([0, 1], [0, 1], color="navy", lw=2, linestyle="--")
# Set limits for x and y axes
ax.set_xlim([0.0, 1.0])
ax.set_ylim([0.0, 1.05])
# Set labels, title, and ticks with specified font sizes
ax.set_xlabel("False Positive Rate", fontsize=14)
ax.set_ylabel("True Positive Rate", fontsize=14)
ax.set_title("Driver Prediction", fontsize=14)
ax.tick_params(axis="both", which="major", labelsize=14)
# Add legend
ax.legend(loc="lower right", fontsize=14)
# Adjust layout to fit the elements within the figure area
plt.tight_layout()
# Save figure if SAVE_FIGURES is True
if SAVE_FIGURES:
save_path = FIG_DIR / DATASET / "ROC_curve.svg"
fig.savefig(save_path, format="svg", transparent=True, bbox_inches="tight")
plt.show()
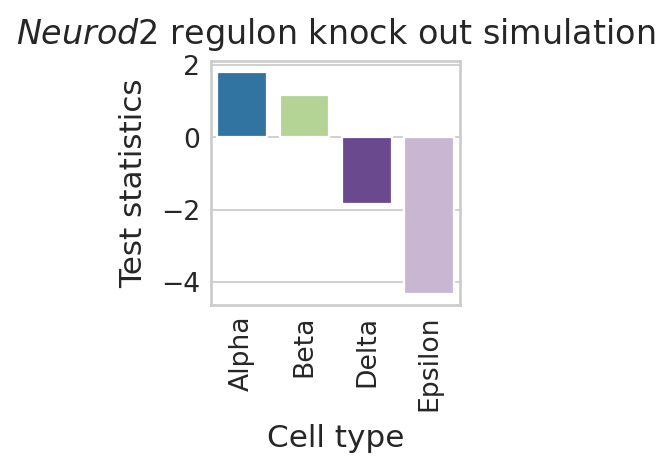
Visualize Neurod2 perturbation#
color_label = "cell_type"
df = pd.DataFrame(regvelo_prediction[0].loc["Neurod2", :])
df.columns = ["Test statistics"]
df["Cell type"] = regvelo_prediction[0].columns.tolist()
order = TERMINAL_STATES
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(2, 2))
sns.barplot(
data=df,
y="Test statistics",
x="Cell type",
palette=palette,
order=order,
ax=ax,
)
# ax.set(ylim=(-0.05, 0.05))
ax.tick_params(axis="x", rotation=90)
plt.title("$\\mathit{" + "Neurod2" + "}$ regulon knock out simulation")
if SAVE_FIGURES:
plt.savefig(
FIG_DIR / DATASET / "Neurod2_perturbation_simulation.svg",
format="svg",
transparent=True,
bbox_inches="tight",
)
# Show the plot
plt.show()
Load original perturbation prediction results to reproduce results in manuscript#
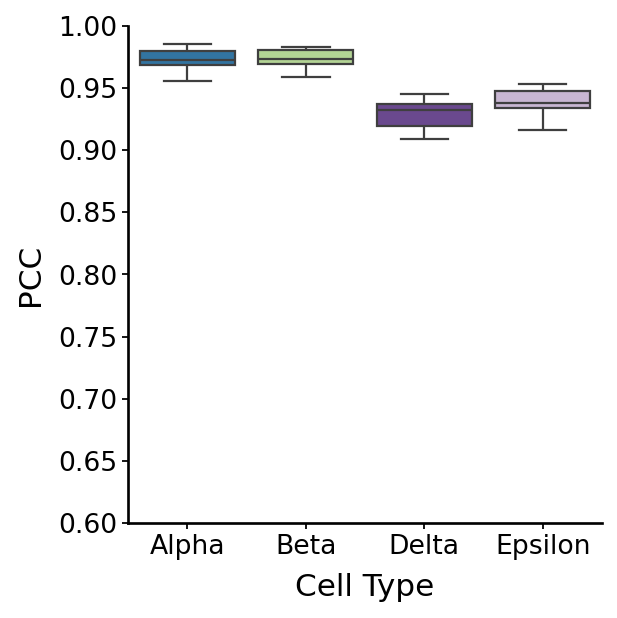
Benchmark perturbation effect estimation robustness#
regvelo_prediction_all = aggregate_model_predictions(
DATA_DIR / DATASET / "raw" / "TF_perturbation", method="t-statistics"
)
for i in range(len(regvelo_prediction_all)):
regvelo_prediction_all[i].columns = regvelo_prediction_all[i].columns + f"_{i}"
rgv_p_all = pd.concat(regvelo_prediction_all, axis=1)
correlation_results = {}
for fate in TERMINAL_STATES:
fate_cols = [f"{fate}_{i}" for i in range(5)]
fate_df = rgv_p_all[fate_cols]
correlation_results[f"{fate}"] = fate_df.corr()
corr_all = []
for fate in TERMINAL_STATES: # Assuming there are 4 metrics
correlation_matrix = correlation_results[f"{fate}"]
corr_pairwise = correlation_matrix.where(np.triu(np.ones(correlation_matrix.shape), k=1).astype(bool))
corr_all = corr_all + corr_pairwise.stack().tolist()
df = pd.DataFrame({"Correlation": corr_all, "Group": np.repeat(TERMINAL_STATES, len(corr_pairwise.stack().tolist()))})
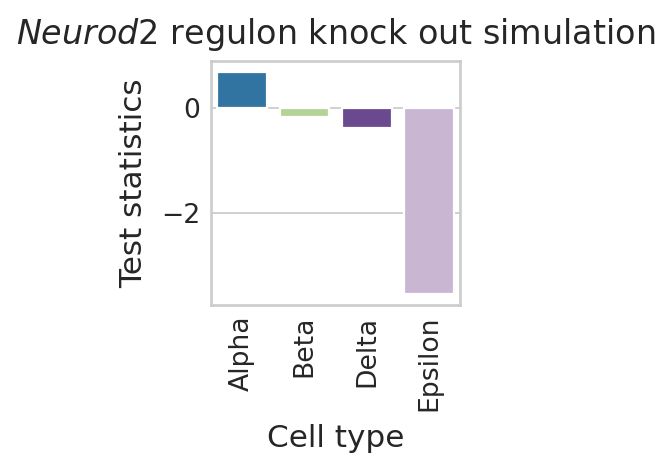
Visualize Neurod2 perturbation effects#
regvelo_prediction_all = aggregate_model_predictions(
DATA_DIR / DATASET / "raw" / "TF_perturbation", method="t-statistics"
)
color_label = "cell_type"
df = pd.DataFrame(regvelo_prediction_all[0].loc["Neurod2", :])
df.columns = ["Test statistics"]
df["Cell type"] = regvelo_prediction_all[0].columns.tolist()
order = TERMINAL_STATES
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(2, 2))
sns.barplot(
data=df,
y="Test statistics",
x="Cell type",
palette=palette,
order=order,
ax=ax,
)
ax.tick_params(axis="x", rotation=90)
plt.title("$\\mathit{" + "Neurod2" + "}$ regulon knock out simulation")
if SAVE_FIGURES:
plt.savefig(
FIG_DIR / DATASET / "Neurod2_perturbation_simulation.svg",
format="svg",
transparent=True,
bbox_inches="tight",
)
# Show the plot
plt.show()
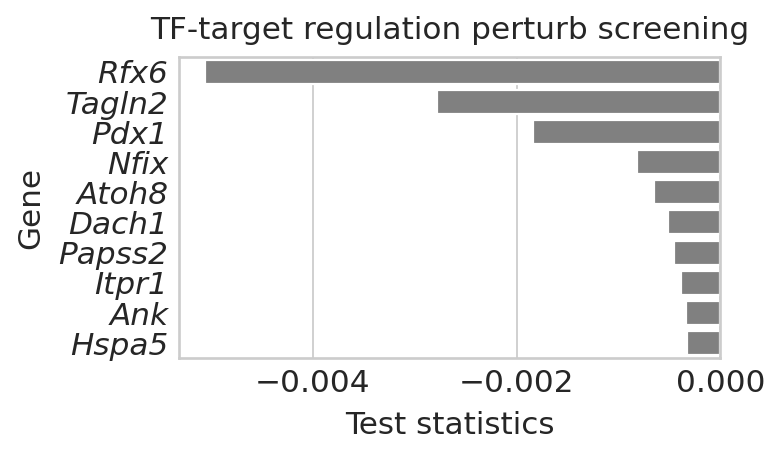
Load target screening results for Neurod2#
Neurod2_target_perturbation = aggregate_model_predictions(
DATA_DIR / DATASET / "raw" / "Neurod2_perturbation", method="t-statistics"
)
## rank the Epsilon prediction results
with mplscience.style_context(): # Entering the custom style context
sns.set_style("whitegrid")
gene_scores = pd.DataFrame(
{
"Gene": Neurod2_target_perturbation[0].index.tolist(),
"Score": Neurod2_target_perturbation[0].loc[:, "Epsilon"],
}
)
gene_scores.loc[:, "weights"] = gene_scores.loc[:, "Score"].abs()
# gene_scores = gene_scores.sort_values(by='weights', ascending=False).iloc[:10,:]
gene_scores = gene_scores.sort_values(by="Score", ascending=True).iloc[:10, :]
# Create the horizontal bar plot using Seaborn
plt.figure(figsize=(5, 3)) # Set the figure size
g = sns.barplot(x="Score", y="Gene", data=gene_scores, color="grey")
# Customize plot aesthetics
g.set_ylabel("Gene", fontsize=14)
g.set_xlabel("Test statistics", fontsize=14)
# Customize tick parameters for better readability
g.tick_params(axis="x", labelsize=14)
g.tick_params(axis="y", labelsize=14)
g.set_title("TF-target regulation perturb screening", fontsize=14)
plt.tight_layout()
plt.setp(g.get_yticklabels(), fontstyle="italic")
if SAVE_FIGURES:
save_path = FIG_DIR / DATASET / "Neurod2_target_screening.svg"
fig.savefig(save_path, format="svg", transparent=True, bbox_inches="tight")
plt.show()
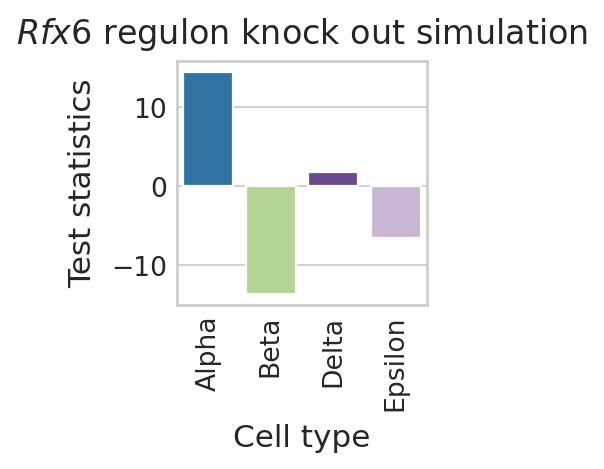
Predict Rfx6 perturbation effects#
color_label = "cell_type"
df = pd.DataFrame(regvelo_prediction_all[0].loc["Rfx6", :])
df.columns = ["Test statistics"]
df["Cell type"] = regvelo_prediction_all[0].columns.tolist()
order = TERMINAL_STATES
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(2, 2))
sns.barplot(
data=df,
y="Test statistics",
x="Cell type",
palette=palette,
order=order,
ax=ax,
)
# ax.set(ylim=(-0.05, 0.05))
ax.tick_params(axis="x", rotation=90)
plt.title("$\\mathit{" + "Rfx6" + "}$ regulon knock out simulation")
if SAVE_FIGURES:
plt.savefig(
FIG_DIR / DATASET / "Rfx6_perturbation_simulation.svg", format="svg", transparent=True, bbox_inches="tight"
)
# Show the plot
plt.show()