Compare estimated velocity robustness between RegVelo and veloVI#
Notebook for evaluating robustness of velocity estimation.
Library imports#
import pandas as pd
import scipy
import matplotlib.pyplot as plt
import mplscience
import seaborn as sns
import scanpy as sc
import scvelo as scv
from rgv_tools import DATA_DIR, FIG_DIR
from rgv_tools.benchmarking import compute_average_correlations
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_csv from `anndata` is deprecated. Import anndata.io.read_csv instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_excel from `anndata` is deprecated. Import anndata.io.read_excel instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_hdf from `anndata` is deprecated. Import anndata.io.read_hdf instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_loom from `anndata` is deprecated. Import anndata.io.read_loom instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_mtx from `anndata` is deprecated. Import anndata.io.read_mtx instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_text from `anndata` is deprecated. Import anndata.io.read_text instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_umi_tools from `anndata` is deprecated. Import anndata.io.read_umi_tools instead.
warnings.warn(msg, FutureWarning)
Constants#
DATASET = "hematopoiesis"
SAVE_FIGURES = True
if SAVE_FIGURES:
(FIG_DIR / DATASET).mkdir(parents=True, exist_ok=True)
Compute estimation robustness#
velo_m = []
time_m = []
for nrun in range(10):
velo_m.append(sc.read_h5ad(DATA_DIR / DATASET / "processed" / f"rgv_adata_runs_{nrun}.h5ad").layers["velocity"])
time_m.append(sc.read_h5ad(DATA_DIR / DATASET / "processed" / f"rgv_adata_runs_{nrun}.h5ad").layers["fit_t"])
velo_rgv = compute_average_correlations(velo_m, method="p")
time_rgv = compute_average_correlations(time_m, method="sp")
velo_m = []
time_m = []
for nrun in range(10):
velo_m.append(sc.read_h5ad(DATA_DIR / DATASET / "processed" / f"vi_adata_runs_{nrun}.h5ad").layers["velocity"])
time_m.append(sc.read_h5ad(DATA_DIR / DATASET / "processed" / f"vi_adata_runs_{nrun}.h5ad").layers["fit_t"])
velo_vi = compute_average_correlations(velo_m, method="p")
time_vi = compute_average_correlations(time_m, method="sp")
Compute velocity confidence#
confi_rgv = []
for nrun in range(10):
adata = sc.read_h5ad(DATA_DIR / DATASET / "processed" / f"rgv_adata_runs_{nrun}.h5ad")
scv.tl.velocity_graph(adata)
scv.tl.velocity_confidence(adata)
confi_rgv.append(adata.obs["velocity_confidence"].mean())
computing velocity graph (using 1/112 cores)
WARNING: Unable to create progress bar. Consider installing `tqdm` as `pip install tqdm` and `ipywidgets` as `pip install ipywidgets`,
or disable the progress bar using `show_progress_bar=False`.
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
--> added 'velocity_length' (adata.obs)
--> added 'velocity_confidence' (adata.obs)
--> added 'velocity_confidence_transition' (adata.obs)
computing velocity graph (using 1/112 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
--> added 'velocity_length' (adata.obs)
--> added 'velocity_confidence' (adata.obs)
--> added 'velocity_confidence_transition' (adata.obs)
computing velocity graph (using 1/112 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
--> added 'velocity_length' (adata.obs)
--> added 'velocity_confidence' (adata.obs)
--> added 'velocity_confidence_transition' (adata.obs)
computing velocity graph (using 1/112 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
--> added 'velocity_length' (adata.obs)
--> added 'velocity_confidence' (adata.obs)
--> added 'velocity_confidence_transition' (adata.obs)
computing velocity graph (using 1/112 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
--> added 'velocity_length' (adata.obs)
--> added 'velocity_confidence' (adata.obs)
--> added 'velocity_confidence_transition' (adata.obs)
computing velocity graph (using 1/112 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
--> added 'velocity_length' (adata.obs)
--> added 'velocity_confidence' (adata.obs)
--> added 'velocity_confidence_transition' (adata.obs)
computing velocity graph (using 1/112 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
--> added 'velocity_length' (adata.obs)
--> added 'velocity_confidence' (adata.obs)
--> added 'velocity_confidence_transition' (adata.obs)
computing velocity graph (using 1/112 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
--> added 'velocity_length' (adata.obs)
--> added 'velocity_confidence' (adata.obs)
--> added 'velocity_confidence_transition' (adata.obs)
computing velocity graph (using 1/112 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
--> added 'velocity_length' (adata.obs)
--> added 'velocity_confidence' (adata.obs)
--> added 'velocity_confidence_transition' (adata.obs)
computing velocity graph (using 1/112 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
--> added 'velocity_length' (adata.obs)
--> added 'velocity_confidence' (adata.obs)
--> added 'velocity_confidence_transition' (adata.obs)
confi_vi = []
for nrun in range(10):
adata = sc.read_h5ad(DATA_DIR / DATASET / "processed" / f"vi_adata_runs_{nrun}.h5ad")
scv.tl.velocity_graph(adata)
scv.tl.velocity_confidence(adata)
confi_vi.append(adata.obs["velocity_confidence"].mean())
computing velocity graph (using 1/112 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
--> added 'velocity_length' (adata.obs)
--> added 'velocity_confidence' (adata.obs)
--> added 'velocity_confidence_transition' (adata.obs)
computing velocity graph (using 1/112 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
--> added 'velocity_length' (adata.obs)
--> added 'velocity_confidence' (adata.obs)
--> added 'velocity_confidence_transition' (adata.obs)
computing velocity graph (using 1/112 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
--> added 'velocity_length' (adata.obs)
--> added 'velocity_confidence' (adata.obs)
--> added 'velocity_confidence_transition' (adata.obs)
computing velocity graph (using 1/112 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
--> added 'velocity_length' (adata.obs)
--> added 'velocity_confidence' (adata.obs)
--> added 'velocity_confidence_transition' (adata.obs)
computing velocity graph (using 1/112 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
--> added 'velocity_length' (adata.obs)
--> added 'velocity_confidence' (adata.obs)
--> added 'velocity_confidence_transition' (adata.obs)
computing velocity graph (using 1/112 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
--> added 'velocity_length' (adata.obs)
--> added 'velocity_confidence' (adata.obs)
--> added 'velocity_confidence_transition' (adata.obs)
computing velocity graph (using 1/112 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
--> added 'velocity_length' (adata.obs)
--> added 'velocity_confidence' (adata.obs)
--> added 'velocity_confidence_transition' (adata.obs)
computing velocity graph (using 1/112 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
--> added 'velocity_length' (adata.obs)
--> added 'velocity_confidence' (adata.obs)
--> added 'velocity_confidence_transition' (adata.obs)
computing velocity graph (using 1/112 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
--> added 'velocity_length' (adata.obs)
--> added 'velocity_confidence' (adata.obs)
--> added 'velocity_confidence_transition' (adata.obs)
computing velocity graph (using 1/112 cores)
finished (0:00:01) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
--> added 'velocity_length' (adata.obs)
--> added 'velocity_confidence' (adata.obs)
--> added 'velocity_confidence_transition' (adata.obs)
Plot benchmark results#
dfs = []
g_df = pd.DataFrame({"velocity correlation": velo_rgv})
g_df["Method"] = "RegVelo"
dfs.append(g_df)
g_df = pd.DataFrame({"velocity correlation": velo_vi})
g_df["Method"] = "veloVI"
dfs.append(g_df)
velo_df = pd.concat(dfs, axis=0)
velo_df["Method"] = velo_df["Method"].astype("category")
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(3, 3))
pal = {"RegVelo": "#0173b2", "veloVI": "#de8f05"}
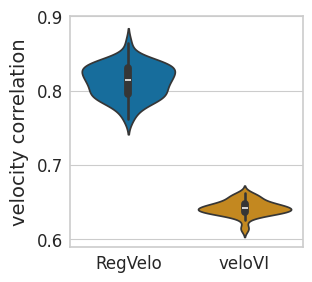
sns.violinplot(
data=velo_df,
ax=ax,
# orient="h",
x="Method",
y="velocity correlation",
order=["RegVelo", "veloVI"],
palette=pal,
)
# plt.legend(title='', loc='lower center', bbox_to_anchor=(0.5, -0.6), ncol=3)
ax.set_yticks([0.6, 0.7, 0.8, 0.9])
ax.set_yticklabels([0.6, 0.7, 0.8, 0.9])
plt.xlabel("")
if SAVE_FIGURES:
fig.savefig(FIG_DIR / DATASET / "velocity_robustness.svg", format="svg", transparent=True, bbox_inches="tight")
plt.show()
dfs = []
g_df = pd.DataFrame({"time correlation": time_rgv})
g_df["Method"] = "RegVelo"
dfs.append(g_df)
g_df = pd.DataFrame({"time correlation": time_vi})
g_df["Method"] = "veloVI"
dfs.append(g_df)
time_df = pd.concat(dfs, axis=0)
time_df["Method"] = time_df["Method"].astype("category")
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(3, 3))
pal = {"RegVelo": "#0173b2", "veloVI": "#de8f05"}
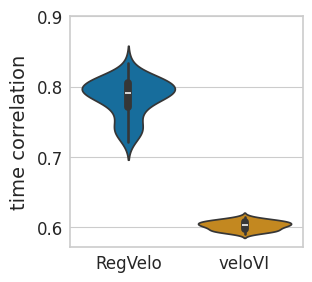
sns.violinplot(
data=time_df,
ax=ax,
# orient="h",
x="Method",
y="time correlation",
order=["RegVelo", "veloVI"],
palette=pal,
)
# plt.legend(title='', loc='lower center', bbox_to_anchor=(0.5, -0.6), ncol=3)
ax.set_yticks([0.6, 0.7, 0.8, 0.9])
ax.set_yticklabels([0.6, 0.7, 0.8, 0.9])
plt.xlabel("")
if SAVE_FIGURES:
fig.savefig(FIG_DIR / DATASET / "time_robustness.svg", format="svg", transparent=True, bbox_inches="tight")
plt.show()
dfs = []
g_df = pd.DataFrame({"velocity confidence": confi_rgv})
g_df["Method"] = "RegVelo"
dfs.append(g_df)
g_df = pd.DataFrame({"velocity confidence": confi_vi})
g_df["Method"] = "veloVI"
dfs.append(g_df)
confi_df = pd.concat(dfs, axis=0)
confi_df["Method"] = confi_df["Method"].astype("category")
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(3, 3))
pal = {"RegVelo": "#0173b2", "veloVI": "#de8f05"}
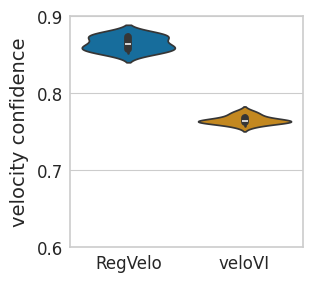
sns.violinplot(
data=confi_df,
ax=ax,
# orient="h",
x="Method",
y="velocity confidence",
order=["RegVelo", "veloVI"],
palette=pal,
)
# plt.legend(title='', loc='lower center', bbox_to_anchor=(0.5, -0.6), ncol=3)
ax.set_yticks([0.6, 0.7, 0.8, 0.9])
ax.set_yticklabels([0.6, 0.7, 0.8, 0.9])
plt.xlabel("")
if SAVE_FIGURES:
fig.savefig(FIG_DIR / DATASET / "velocity_confidence.svg", format="svg", transparent=True, bbox_inches="tight")
plt.show()
Test significance#
scipy.stats.ttest_ind(velo_rgv, velo_vi, alternative="greater")
TtestResult(statistic=47.5942306011296, pvalue=6.913291028470966e-65, df=88.0)
scipy.stats.ttest_ind(time_rgv, time_vi, alternative="greater")
TtestResult(statistic=44.393343875292615, pvalue=2.484323625884492e-62, df=88.0)
scipy.stats.ttest_ind(confi_rgv, confi_vi, alternative="greater")
TtestResult(statistic=29.233032070958988, pvalue=6.332341886443939e-17, df=18.0)