Comparing lineage-associated gene rankings#
Notebook compares the performance of different methods for ranking lineage-associated genes.
Library imports#
import pandas as pd
import matplotlib.pyplot as plt
import scvelo as scv
from rgv_tools import DATA_DIR, FIG_DIR
from rgv_tools.benchmarking import (
get_aucs,
get_gene_ranks,
get_optimal_auc,
get_rank_threshold,
get_var_ranks,
plot_gene_ranking,
)
from rgv_tools.core import METHOD_PALETTE_RANKING
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_csv from `anndata` is deprecated. Import anndata.io.read_csv instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_excel from `anndata` is deprecated. Import anndata.io.read_excel instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_hdf from `anndata` is deprecated. Import anndata.io.read_hdf instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_loom from `anndata` is deprecated. Import anndata.io.read_loom instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_mtx from `anndata` is deprecated. Import anndata.io.read_mtx instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_text from `anndata` is deprecated. Import anndata.io.read_text instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/dynamo/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_umi_tools from `anndata` is deprecated. Import anndata.io.read_umi_tools instead.
warnings.warn(msg, FutureWarning)
General settings#
plt.rcParams["svg.fonttype"] = "none"
scv.settings.set_figure_params("scvelo", dpi_save=400, dpi=80, transparent=True, fontsize=14, color_map="viridis")
Constants#
DATASET = "hematopoiesis"
SAVE_FIGURES = True
if SAVE_FIGURES:
(FIG_DIR / DATASET).mkdir(parents=True, exist_ok=True)
FIGURE_FORMAT = "svg"
TERMINAL_STATES = ["Ery", "Mon"]
Data loading#
drivers_rgv = pd.read_csv(DATA_DIR / DATASET / "results" / "rgv_ranking.csv", index_col=0)
drivers_scv = pd.read_csv(DATA_DIR / DATASET / "results" / "scv_ranking.csv", index_col=0)
drivers_vi = pd.read_csv(DATA_DIR / DATASET / "results" / "vi_ranking.csv", index_col=0)
Load ground truth
Ery_g = pd.read_csv(DATA_DIR / DATASET / "raw" / "Ery_lineage_associated_genes.csv", index_col=0).iloc[:, 0]
Mon_g = pd.read_csv(DATA_DIR / DATASET / "raw" / "Mon_lineage_associated_genes.csv", index_col=0).iloc[:, 0]
Ranking#
terminal_states = ["Ery", "Mon"]
for terminal_state in TERMINAL_STATES:
drivers_rgv = drivers_rgv.merge(
pd.DataFrame(drivers_rgv.sort_values(by=f"{terminal_state}_corr", ascending=False).index)
.reset_index()
.rename(columns={"index": f"Corr. rank - {terminal_state}"})
.set_index(0),
left_index=True,
right_index=True,
)
drivers_scv = drivers_scv.merge(
pd.DataFrame(drivers_scv.sort_values(by=f"{terminal_state}_corr", ascending=False).index)
.reset_index()
.rename(columns={"index": f"Corr. rank - {terminal_state}"})
.set_index(0),
left_index=True,
right_index=True,
)
drivers_vi = drivers_vi.merge(
pd.DataFrame(drivers_vi.sort_values(by=f"{terminal_state}_corr", ascending=False).index)
.reset_index()
.rename(columns={"index": f"Corr. rank - {terminal_state}"})
.set_index(0),
left_index=True,
right_index=True,
)
Ery_g = list(set(Ery_g).intersection(drivers_scv.index.tolist()))
Mon_g = list(set(Mon_g).intersection(drivers_scv.index.tolist()))
gene_ranks_rgv = {terminal_state: pd.DataFrame() for terminal_state in TERMINAL_STATES}
## load Ery
_df = get_var_ranks(var_names=Ery_g, drivers=drivers_rgv, macrostate="Ery", var_type="Driver", model="RegVelo")
gene_ranks_rgv["Ery"] = pd.concat([gene_ranks_rgv["Ery"], _df])
## load Mon
_df = get_var_ranks(var_names=Mon_g, drivers=drivers_rgv, macrostate="Mon", var_type="Driver", model="RegVelo")
gene_ranks_rgv["Mon"] = pd.concat([gene_ranks_rgv["Mon"], _df])
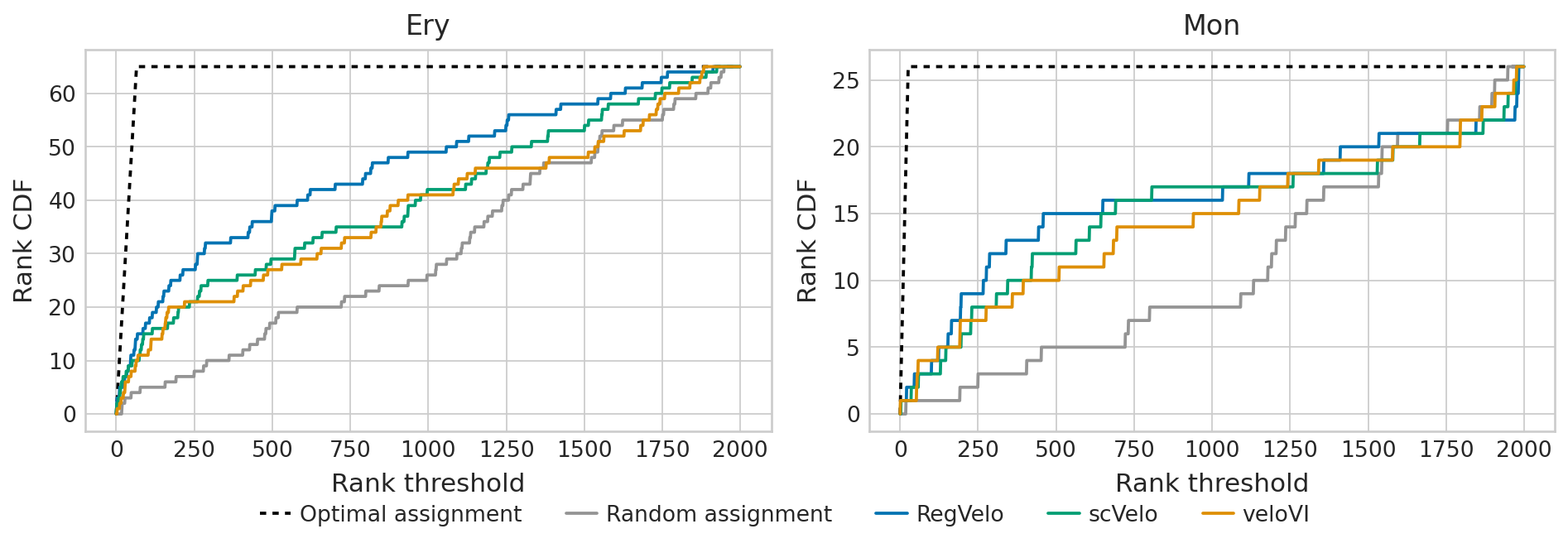
Driver towards Ery for RegVelo in top 100: 17 (out of 65)
Driver towards Mon for RegVelo in top 100: 4 (out of 26)
for terminal_state in gene_ranks_rgv.keys():
# Sort the DataFrame for each terminal state
gene_ranks_rgv[terminal_state] = gene_ranks_rgv[terminal_state].sort_values(f"Corr. rank - {terminal_state}")
# Create the path for the output file
output_path = DATA_DIR / DATASET / "results" / f"gene_ranks_{terminal_state}-rgvelo.csv"
# Save the sorted DataFrame to a CSV file, one for each terminal state
gene_ranks_rgv[terminal_state].to_csv(output_path)
gene_ranks_scv = {terminal_state: pd.DataFrame() for terminal_state in TERMINAL_STATES}
## load Ery
_df = get_var_ranks(var_names=Ery_g, drivers=drivers_scv, macrostate="Ery", var_type="Driver", model="scVelo")
gene_ranks_scv["Ery"] = pd.concat([gene_ranks_scv["Ery"], _df])
## load Mon
_df = get_var_ranks(var_names=Mon_g, drivers=drivers_scv, macrostate="Mon", var_type="Driver", model="scVelo")
gene_ranks_scv["Mon"] = pd.concat([gene_ranks_scv["Mon"], _df])
Driver towards Ery for scVelo in top 100: 15 (out of 65)
Driver towards Mon for scVelo in top 100: 3 (out of 26)
for terminal_state in gene_ranks_scv.keys():
# Sort the DataFrame for each terminal state
gene_ranks_scv[terminal_state] = gene_ranks_scv[terminal_state].sort_values(f"Corr. rank - {terminal_state}")
# Create the path for the output file
output_path = DATA_DIR / DATASET / "results" / f"gene_ranks_{terminal_state}-scVelo.csv"
# Save the sorted DataFrame to a CSV file, one for each terminal state
gene_ranks_scv[terminal_state].to_csv(output_path)
gene_ranks_vi = {terminal_state: pd.DataFrame() for terminal_state in TERMINAL_STATES}
## load Ery
_df = get_var_ranks(var_names=Ery_g, drivers=drivers_vi, macrostate="Ery", var_type="Driver", model="veloVI")
gene_ranks_vi["Ery"] = pd.concat([gene_ranks_vi["Ery"], _df])
## load Mon
_df = get_var_ranks(var_names=Mon_g, drivers=drivers_vi, macrostate="Mon", var_type="Driver", model="veloVI")
gene_ranks_vi["Mon"] = pd.concat([gene_ranks_vi["Mon"], _df])
Driver towards Ery for veloVI in top 100: 11 (out of 65)
Driver towards Mon for veloVI in top 100: 4 (out of 26)
for terminal_state in gene_ranks_vi.keys():
# Sort the DataFrame for each terminal state
gene_ranks_vi[terminal_state] = gene_ranks_vi[terminal_state].sort_values(f"Corr. rank - {terminal_state}")
# Create the path for the output file
output_path = DATA_DIR / DATASET / "results" / f"gene_ranks_{terminal_state}-veloVI.csv"
# Save the sorted DataFrame to a CSV file, one for each terminal state
gene_ranks_vi[terminal_state].to_csv(output_path)
gene_ranks = get_gene_ranks(TERMINAL_STATES, DATA_DIR, DATASET)
methods = {}
n_methods = {}
for terminal_state in TERMINAL_STATES:
methods[terminal_state] = gene_ranks[terminal_state]["Algorithm"].unique().tolist()
if "Run" in gene_ranks[terminal_state].columns:
n_methods[terminal_state] = gene_ranks[terminal_state][["Algorithm", "Run"]].drop_duplicates().shape[0]
else:
n_methods[terminal_state] = len(methods[terminal_state])
dfs = get_rank_threshold(gene_ranks=gene_ranks, n_methods=n_methods, TERMINAL_STATES=TERMINAL_STATES)
pal = METHOD_PALETTE_RANKING
plot_gene_ranking(
rank_threshold=dfs,
methods=methods,
palette=pal,
TERMINAL_STATES=TERMINAL_STATES,
path=FIG_DIR / DATASET / f"Gene Ranking.{FIGURE_FORMAT}",
format=FIGURE_FORMAT,
)
optimal_aucs = {}
for terminal_state in TERMINAL_STATES:
if "Run" in gene_ranks[terminal_state].columns:
optimal_aucs[terminal_state] = gene_ranks[terminal_state].groupby(["Algorithm"]).size() / gene_ranks[
terminal_state
].groupby(["Algorithm"]).apply(lambda x: len(x["Run"].unique()))
else:
optimal_aucs[terminal_state] = gene_ranks[terminal_state].groupby("Algorithm").size()
optimal_aucs[terminal_state] = get_optimal_auc(optimal_aucs[terminal_state])
_, auc_rel_df = get_aucs(
gene_ranking_dfs=dfs, optimal_aucs=optimal_aucs, methods=methods, TERMINAL_STATES=TERMINAL_STATES
)
auc_rel_df
| Ery | Mon | |
|---|---|---|
| scVelo | 0.634002 | 0.588447 |
| Optimal assignment | 0.999238 | 0.999245 |
| Random assignment | 0.490443 | 0.423125 |
| RegVelo | 0.724887 | 0.620126 |
| veloVI | 0.604311 | 0.562961 |