Benchmark cell fate and velocity identifiability#
Notebook for evaluate cellular velocity and cell fate identifiability
Library imports#
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import mplscience
import seaborn as sns
import cellrank as cr
import scanpy as sc
import scvelo as scv
from regvelo import REGVELOVI
from rgv_tools import DATA_DIR, FIG_DIR
from rgv_tools.benchmarking import set_output
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_csv from `anndata` is deprecated. Import anndata.io.read_csv instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_excel from `anndata` is deprecated. Import anndata.io.read_excel instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_hdf from `anndata` is deprecated. Import anndata.io.read_hdf instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_loom from `anndata` is deprecated. Import anndata.io.read_loom instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_mtx from `anndata` is deprecated. Import anndata.io.read_mtx instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_text from `anndata` is deprecated. Import anndata.io.read_text instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_umi_tools from `anndata` is deprecated. Import anndata.io.read_umi_tools instead.
warnings.warn(msg, FutureWarning)
General settings#
%matplotlib inline
plt.rcParams["svg.fonttype"] = "none"
sns.reset_defaults()
sns.reset_orig()
scv.settings.set_figure_params("scvelo", dpi_save=400, dpi=80, transparent=True, fontsize=14, color_map="viridis")
Constants#
DATASET = "pancreatic_endocrinogenesis"
SAVE_DATA = True
if SAVE_DATA:
(DATA_DIR / DATASET / "results").mkdir(parents=True, exist_ok=True)
SAVE_FIGURES = True
if SAVE_FIGURES:
(FIG_DIR / DATASET).mkdir(parents=True, exist_ok=True)
TERMINAL_STATES = ["Alpha", "Beta", "Delta", "Epsilon"]
Function defination#
def cal_cell_fate(adata, terminal_states, cluster_key, n_states=7):
"""Cellrank's typical analysis pipeline."""
vk = cr.kernels.VelocityKernel(adata).compute_transition_matrix()
ck = cr.kernels.ConnectivityKernel(adata).compute_transition_matrix()
combined_kernel = 0.8 * vk + 0.2 * ck
estimator = cr.estimators.GPCCA(combined_kernel)
## evaluate the fate prob on original space
estimator.compute_macrostates(n_states=n_states, n_cells=30, cluster_key=cluster_key)
## set a high number of states, and merge some of them and rename
macrostate = estimator.macrostates.cat.categories.tolist()
terminal_states_recovered = list(set(macrostate).intersection(terminal_states))
## check if all terminal state is coverred
if len(set(terminal_states_recovered).intersection(terminal_states)) != len(terminal_states):
ValueError("Terminal states are not recoverred in perturbation, please increase the number of the states!")
estimator.set_terminal_states(terminal_states)
estimator.compute_fate_probabilities(solver="direct")
fb = estimator.fate_probabilities
sampleID = adata.obs.index.tolist()
fate_name = fb.names.tolist()
fb = pd.DataFrame(fb, index=sampleID, columns=fate_name)
return fb
Data loading#
adata = sc.read_h5ad(DATA_DIR / DATASET / "processed" / "adata_preprocessed.h5ad")
palette = dict(zip(adata.obs["clusters"].cat.categories, adata.uns["clusters_colors"]))
Model loading#
adata_m = []
velo_m = []
for nrun in range(0, 5):
model = DATA_DIR / DATASET / "processed" / "perturb_repeat_runs" / f"rgv_model_{nrun}"
### load model
vae = REGVELOVI.load(model, adata)
# grn1 = reg_vae.module.v_encoder.fc1.weight.data.cpu().detach().clone()
set_output(adata, vae, n_samples=30, batch_size=adata.n_obs)
adata_m.append(adata.copy())
velo_m.append(adata.layers["velocity"])
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:204: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_0/model.pt already downloaded
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_1/model.pt already downloaded
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:204: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_2/model.pt already downloaded
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:204: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_3/model.pt already downloaded
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:204: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
INFO File
/ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/perturb_repeat_runs/rgv_
model_4/model.pt already downloaded
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:204: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
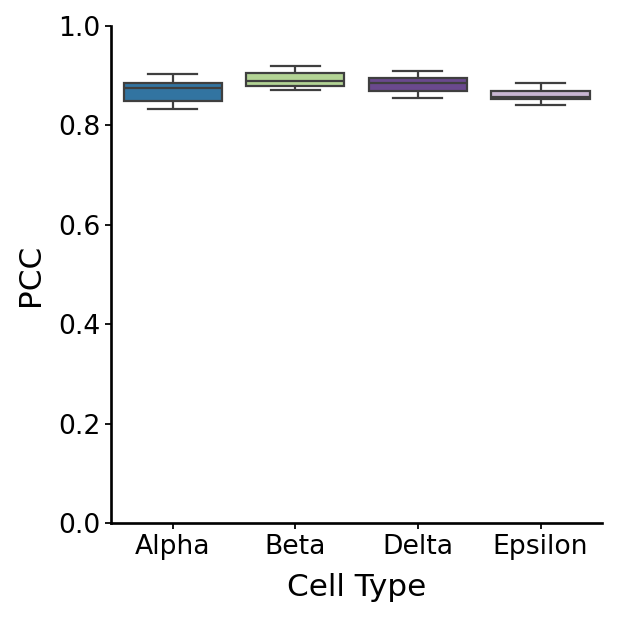
Cellular velocity#
velo_arrays = np.stack(velo_m, axis=2)
N = adata.shape[0]
# Calculate pairwise cell correlation among two arrays
correlations = []
for i in range(velo_arrays.shape[2] + 1):
for j in range(i + 1, (velo_arrays.shape[2])):
if i != j:
correlation_matrix = np.corrcoef(velo_arrays[:, :, i], velo_arrays[:, :, j])
# Extract upper triangle part (excluding diagonal)
correlation_matrix = correlation_matrix[0:N, N : (2 * N)]
corr = np.diag(correlation_matrix)
corr = [
np.mean(np.array(corr)[adata.obs["clusters"] == "Alpha"]),
np.mean(np.array(corr)[adata.obs["clusters"] == "Beta"]),
np.mean(np.array(corr)[adata.obs["clusters"] == "Delta"]),
np.mean(np.array(corr)[adata.obs["clusters"] == "Epsilon"]),
]
correlations.append(corr)
correlations = np.array(correlations)
df = pd.DataFrame(
{
"Correlation": correlations[:, 0].tolist()
+ correlations[:, 1].tolist()
+ correlations[:, 2].tolist()
+ correlations[:, 3].tolist(),
"Group": np.repeat(TERMINAL_STATES, correlations.shape[0]),
}
)
with mplscience.style_context():
fig, ax = plt.subplots(figsize=(4, 4))
sns.boxplot(data=df, x="Group", y="Correlation", palette=palette, ax=ax)
plt.xlabel("Cell Type")
plt.ylabel("PCC")
plt.ylim(0, 1)
plt.tight_layout()
plt.show()
df_rgv_velo = df.copy()
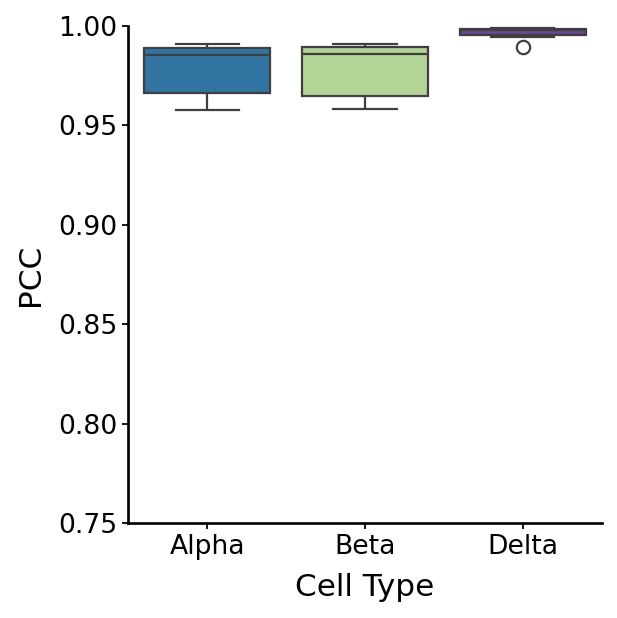
Cell fate#
fate_list = []
for n, ad in enumerate(adata_m):
fate = cal_cell_fate(ad, terminal_states=TERMINAL_STATES[:3], cluster_key="clusters", n_states=8)
fate.columns = fate.columns + f"_{n}"
fate_list.append(fate)
[0]PETSC ERROR: ------------------------------------------------------------------------
[0]PETSC ERROR: Caught signal number 13 Broken Pipe: Likely while reading or writing to a socket
[0]PETSC ERROR: Try option -start_in_debugger or -on_error_attach_debugger
[0]PETSC ERROR: or see https://petsc.org/release/faq/#valgrind and https://petsc.org/release/faq/
[0]PETSC ERROR: configure using --with-debugging=yes, recompile, link, and run
[0]PETSC ERROR: to get more information on the crash.
Abort(59) on node 0 (rank 0 in comm 0): application called MPI_Abort(MPI_COMM_WORLD, 59) - process 0
[0]PETSC ERROR: ------------------------------------------------------------------------
[0]PETSC ERROR: Caught signal number 13 Broken Pipe: Likely while reading or writing to a socket
[0]PETSC ERROR: Try option -start_in_debugger or -on_error_attach_debugger
[0]PETSC ERROR: or see https://petsc.org/release/faq/#valgrind and https://petsc.org/release/faq/
[0]PETSC ERROR: configure using --with-debugging=yes, recompile, link, and run
[0]PETSC ERROR: to get more information on the crash.
Abort(59) on node 0 (rank 0 in comm 0): application called MPI_Abort(MPI_COMM_WORLD, 59) - process 0
[0]PETSC ERROR: ------------------------------------------------------------------------
[0]PETSC ERROR: Caught signal number 13 Broken Pipe: Likely while reading or writing to a socket
[0]PETSC ERROR: Try option -start_in_debugger or -on_error_attach_debugger
[0]PETSC ERROR: or see https://petsc.org/release/faq/#valgrind and https://petsc.org/release/faq/
[0]PETSC ERROR: configure using --with-debugging=yes, recompile, link, and run
[0]PETSC ERROR: to get more information on the crash.
Abort(59) on node 0 (rank 0 in comm 0): application called MPI_Abort(MPI_COMM_WORLD, 59) - process 0
[0]PETSC ERROR: ------------------------------------------------------------------------
[0]PETSC ERROR: Caught signal number 13 Broken Pipe: Likely while reading or writing to a socket
[0]PETSC ERROR: Try option -start_in_debugger or -on_error_attach_debugger
[0]PETSC ERROR: or see https://petsc.org/release/faq/#valgrind and https://petsc.org/release/faq/
[0]PETSC ERROR: configure using --with-debugging=yes, recompile, link, and run
[0]PETSC ERROR: to get more information on the crash.
Abort(59) on node 0 (rank 0 in comm 0): application called MPI_Abort(MPI_COMM_WORLD, 59) - process 0
[0]PETSC ERROR: ------------------------------------------------------------------------
[0]PETSC ERROR: Caught signal number 13 Broken Pipe: Likely while reading or writing to a socket
[0]PETSC ERROR: Try option -start_in_debugger or -on_error_attach_debugger
[0]PETSC ERROR: or see https://petsc.org/release/faq/#valgrind and https://petsc.org/release/faq/
[0]PETSC ERROR: configure using --with-debugging=yes, recompile, link, and run
[0]PETSC ERROR: to get more information on the crash.
Abort(59) on node 0 (rank 0 in comm 0): application called MPI_Abort(MPI_COMM_WORLD, 59) - process 0
[0]PETSC ERROR: ------------------------------------------------------------------------
[0]PETSC ERROR: Caught signal number 13 Broken Pipe: Likely while reading or writing to a socket
[0]PETSC ERROR: Try option -start_in_debugger or -on_error_attach_debugger
[0]PETSC ERROR: or see https://petsc.org/release/faq/#valgrind and https://petsc.org/release/faq/
[0]PETSC ERROR: configure using --with-debugging=yes, recompile, link, and run
[0]PETSC ERROR: to get more information on the crash.
Abort(59) on node 0 (rank 0 in comm 0): application called MPI_Abort(MPI_COMM_WORLD, 59) - process 0
[0]PETSC ERROR: ------------------------------------------------------------------------
[0]PETSC ERROR: Caught signal number 13 Broken Pipe: Likely while reading or writing to a socket
[0]PETSC ERROR: Try option -start_in_debugger or -on_error_attach_debugger
[0]PETSC ERROR: or see https://petsc.org/release/faq/#valgrind and https://petsc.org/release/faq/
[0]PETSC ERROR: configure using --with-debugging=yes, recompile, link, and run
[0]PETSC ERROR: to get more information on the crash.
Abort(59) on node 0 (rank 0 in comm 0): application called MPI_Abort(MPI_COMM_WORLD, 59) - process 0
[0]PETSC ERROR: ------------------------------------------------------------------------
[0]PETSC ERROR: Caught signal number 13 Broken Pipe: Likely while reading or writing to a socket
[0]PETSC ERROR: Try option -start_in_debugger or -on_error_attach_debugger
[0]PETSC ERROR: or see https://petsc.org/release/faq/#valgrind and https://petsc.org/release/faq/
[0]PETSC ERROR: configure using --with-debugging=yes, recompile, link, and run
[0]PETSC ERROR: to get more information on the crash.
Abort(59) on node 0 (rank 0 in comm 0): application called MPI_Abort(MPI_COMM_WORLD, 59) - process 0
fate_all = pd.concat(fate_list, axis=1)
correlation_results = {}
for fate in TERMINAL_STATES[:3]:
fate_cols = [f"{fate}_{i}" for i in range(5)]
fate_df = fate_all[fate_cols]
correlation_results[f"{fate}"] = fate_df.corr()
corr_all = []
for fate in TERMINAL_STATES[:3]: # Assuming there are 4 metrics
correlation_matrix = correlation_results[f"{fate}"]
corr_pairwise = correlation_matrix.where(np.triu(np.ones(correlation_matrix.shape), k=1).astype(bool))
corr_all = corr_all + corr_pairwise.stack().tolist()
df = pd.DataFrame(
{"Correlation": corr_all, "Group": np.repeat(TERMINAL_STATES[:3], len(corr_pairwise.stack().tolist()))}
)
with mplscience.style_context():
fig, ax = plt.subplots(figsize=(4, 4))
sns.boxplot(data=df, x="Group", y="Correlation", palette=palette, ax=ax)
plt.xlabel("Cell Type")
plt.ylabel("PCC")
plt.ylim(0.75, 1)
plt.tight_layout()
plt.show()
df_rgv_fb = df.copy()
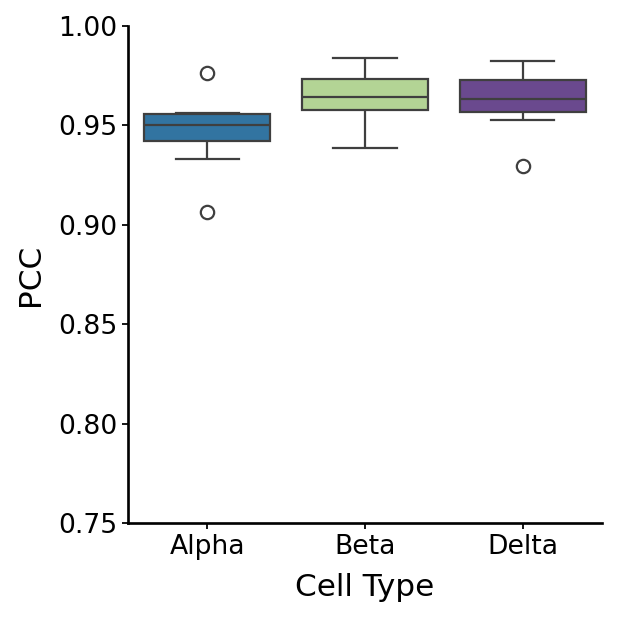
Using veloVI as cell fate analysis baseline#
fate_list = []
for n in range(5):
ad = sc.read_h5ad(DATA_DIR / DATASET / "processed" / f"adata_velovi_run{n}")
fate = cal_cell_fate(ad, terminal_states=TERMINAL_STATES[:3], cluster_key="clusters", n_states=10)
fate.columns = fate.columns + f"_{n}"
fate_list.append(fate)
[0]PETSC ERROR: ------------------------------------------------------------------------
[0]PETSC ERROR: Caught signal number 13 Broken Pipe: Likely while reading or writing to a socket
[0]PETSC ERROR: Try option -start_in_debugger or -on_error_attach_debugger
[0]PETSC ERROR: or see https://petsc.org/release/faq/#valgrind and https://petsc.org/release/faq/
[0]PETSC ERROR: configure using --with-debugging=yes, recompile, link, and run
WARNING: Using `10` components would split a block of complex conjugate eigenvalues. Using `n_components=11`
[0]PETSC ERROR:
WARNING: Unable to compute macrostates with `n_states=10` because it will split complex conjugate eigenvalues. Using `n_states=11`
WARNING: Using `10` components would split a block of complex conjugate eigenvalues. Using `n_components=11`
WARNING: Unable to compute macrostates with `n_states=10` because it will split complex conjugate eigenvalues. Using `n_states=11`
WARNING: Using `10` components would split a block of complex conjugate eigenvalues. Using `n_components=11`
[0]PETSC ERROR:
WARNING: Unable to compute macrostates with `n_states=10` because it will split complex conjugate eigenvalues. Using `n_states=11`
[0]PETSC ERROR:
[0]PETSC ERROR:
WARNING: The following terminal states have different number of cells than requested (30): {'Ductal_2': 26}
fate_all = pd.concat(fate_list, axis=1)
correlation_results = {}
for fate in TERMINAL_STATES[:3]:
fate_cols = [f"{fate}_{i}" for i in range(5)]
fate_df = fate_all[fate_cols]
correlation_results[f"{fate}"] = fate_df.corr()
corr_all = []
for fate in TERMINAL_STATES[:3]: # Assuming there are 4 metrics
correlation_matrix = correlation_results[f"{fate}"]
corr_pairwise = correlation_matrix.where(np.triu(np.ones(correlation_matrix.shape), k=1).astype(bool))
corr_all = corr_all + corr_pairwise.stack().tolist()
df = pd.DataFrame(
{"Correlation": corr_all, "Group": np.repeat(TERMINAL_STATES[:3], len(corr_pairwise.stack().tolist()))}
)
with mplscience.style_context():
fig, ax = plt.subplots(figsize=(4, 4))
sns.boxplot(data=df, x="Group", y="Correlation", palette=palette, ax=ax)
plt.xlabel("Cell Type")
plt.ylabel("PCC")
plt.ylim(0.75, 1)
plt.tight_layout()
plt.show()
df_vi_fb = df.copy()
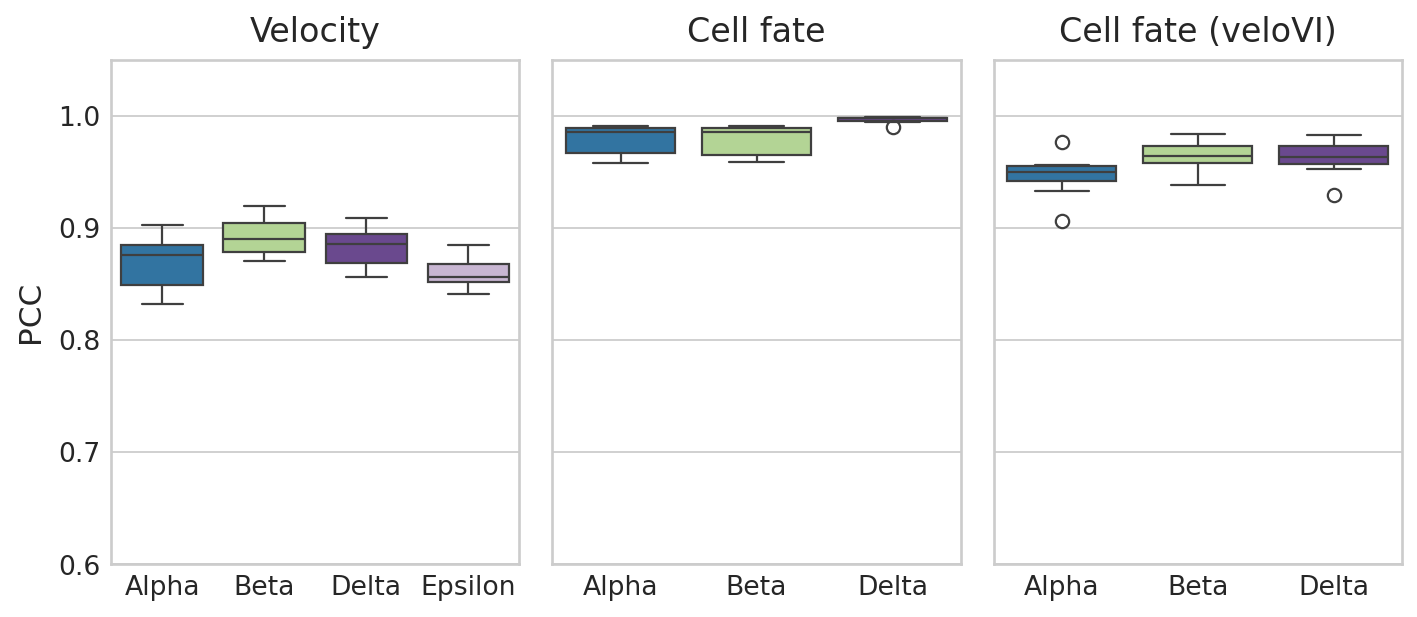
# Create figure with two subplots
with mplscience.style_context(): # Entering the custom style context
sns.set_style("whitegrid")
fig, axes = plt.subplots(1, 3, figsize=(9, 4), sharey=True) # 1 row, 2 columns
# Loop over the subplots and plot data for each run
ax = axes[0]
sns.boxplot(y="Correlation", x="Group", data=df_rgv_velo, ax=ax, palette=palette)
ax.set_title("Velocity")
ax.set_xlabel("")
ax.set_ylabel("PCC")
ax = axes[1]
sns.boxplot(y="Correlation", x="Group", data=df_rgv_fb, ax=ax, palette=palette)
ax.set_title("Cell fate")
ax.set_xlabel("")
ax.set_ylabel("")
ax = axes[2]
sns.boxplot(y="Correlation", x="Group", data=df_vi_fb, ax=ax, palette=palette)
ax.set_title("Cell fate (veloVI)")
ax.set_xlabel("")
ax.set_ylabel("")
# Set a common ylabel
axes[0].set_ylabel("PCC")
plt.ylim(0.6, 1.05)
# Show the plot
plt.tight_layout()
if SAVE_FIGURES:
save_path = FIG_DIR / DATASET / "Identifiability_Benchmark.svg"
fig.savefig(save_path, format="svg", transparent=True, bbox_inches="tight")
plt.show()
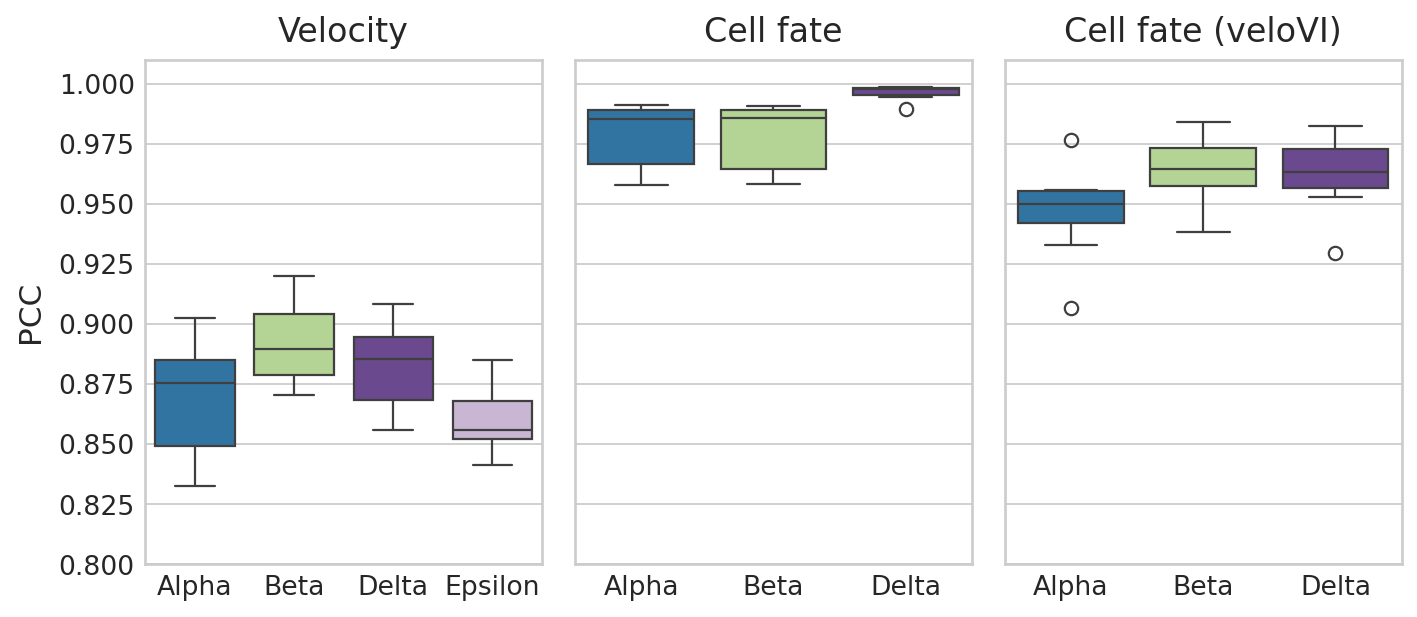
# Create figure with two subplots
with mplscience.style_context(): # Entering the custom style context
sns.set_style("whitegrid")
fig, axes = plt.subplots(1, 3, figsize=(9, 4), sharey=True) # 1 row, 2 columns
# Loop over the subplots and plot data for each run
ax = axes[0]
sns.boxplot(y="Correlation", x="Group", data=df_rgv_velo, ax=ax, palette=palette)
ax.set_title("Velocity")
ax.set_xlabel("")
ax.set_ylabel("PCC")
ax = axes[1]
sns.boxplot(y="Correlation", x="Group", data=df_rgv_fb, ax=ax, palette=palette)
ax.set_title("Cell fate")
ax.set_xlabel("")
ax.set_ylabel("")
ax = axes[2]
sns.boxplot(y="Correlation", x="Group", data=df_vi_fb, ax=ax, palette=palette)
ax.set_title("Cell fate (veloVI)")
ax.set_xlabel("")
ax.set_ylabel("")
# Set a common ylabel
axes[0].set_ylabel("PCC")
plt.ylim(0.8, 1.01)
# Show the plot
plt.tight_layout()
if SAVE_FIGURES:
save_path = FIG_DIR / DATASET / "Identifiability_Benchmark.svg"
fig.savefig(save_path, format="svg", transparent=True, bbox_inches="tight")
plt.show()