Regulation is important for terminal state identification in pancreatic endocrine#
Test the gene regulation roles in predicting terminal states
Library imports#
import random
import pandas as pd
import torch
import matplotlib.pyplot as plt
import mplscience
import seaborn as sns
import cellrank as cr
import scanpy as sc
import scvelo as scv
from regvelo import REGVELOVI
from rgv_tools import DATA_DIR, FIG_DIR
from rgv_tools.benchmarking import set_output
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_csv from `anndata` is deprecated. Import anndata.io.read_csv instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_excel from `anndata` is deprecated. Import anndata.io.read_excel instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_hdf from `anndata` is deprecated. Import anndata.io.read_hdf instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_loom from `anndata` is deprecated. Import anndata.io.read_loom instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_mtx from `anndata` is deprecated. Import anndata.io.read_mtx instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_text from `anndata` is deprecated. Import anndata.io.read_text instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_umi_tools from `anndata` is deprecated. Import anndata.io.read_umi_tools instead.
warnings.warn(msg, FutureWarning)
General settings#
plt.rcParams["svg.fonttype"] = "none"
sns.reset_defaults()
sns.reset_orig()
scv.settings.set_figure_params("scvelo", dpi_save=400, dpi=80, transparent=True, fontsize=14, color_map="viridis")
Constants#
DATASET = "pancreatic_endocrinogenesis"
SAVE_FIGURES = True
if SAVE_FIGURES:
(FIG_DIR / DATASET).mkdir(parents=True, exist_ok=True)
TERMINAL_STATES = ["Beta", "Alpha", "Delta", "Epsilon"]
N_STATES = 7
Data loading#
adata = sc.read_h5ad(DATA_DIR / DATASET / "processed" / "adata_preprocessed.h5ad")
Model loading#
vae = REGVELOVI.load(DATA_DIR / DATASET / "processed" / "rgv_model", adata)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:204: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/rgv_model/model.pt
already downloaded
Delete regulation#
adata_no_regulation = adata.copy()
vae.module.v_encoder.fc1.weight.data = vae.module.v_encoder.fc1.weight.data * 0
set_output(adata_no_regulation, vae, n_samples=30, batch_size=adata.n_obs)
scv.tl.velocity_graph(adata_no_regulation)
computing velocity graph (using 1/112 cores)
finished (0:00:09) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
with mplscience.style_context():
fig, ax = plt.subplots(figsize=(4, 3))
scv.pl.velocity_embedding_stream(adata_no_regulation, basis="umap", title="", legend_loc=False, ax=ax)
if SAVE_FIGURES:
fig.savefig(
FIG_DIR / DATASET / "no_regulation_vector_field.svg", format="svg", transparent=True, bbox_inches="tight"
)
Predict terminal states#
vk = cr.kernels.VelocityKernel(adata_no_regulation)
vk.compute_transition_matrix()
ck = cr.kernels.ConnectivityKernel(adata_no_regulation).compute_transition_matrix()
estimator = cr.estimators.GPCCA(0.8 * vk + 0.2 * ck)
## evaluate the fate prob on original space
estimator.compute_macrostates(n_states=N_STATES, cluster_key="clusters")
GPCCA[kernel=(0.8 * VelocityKernel[n=3696] + 0.2 * ConnectivityKernel[n=3696]), initial_states=None, terminal_states=None]
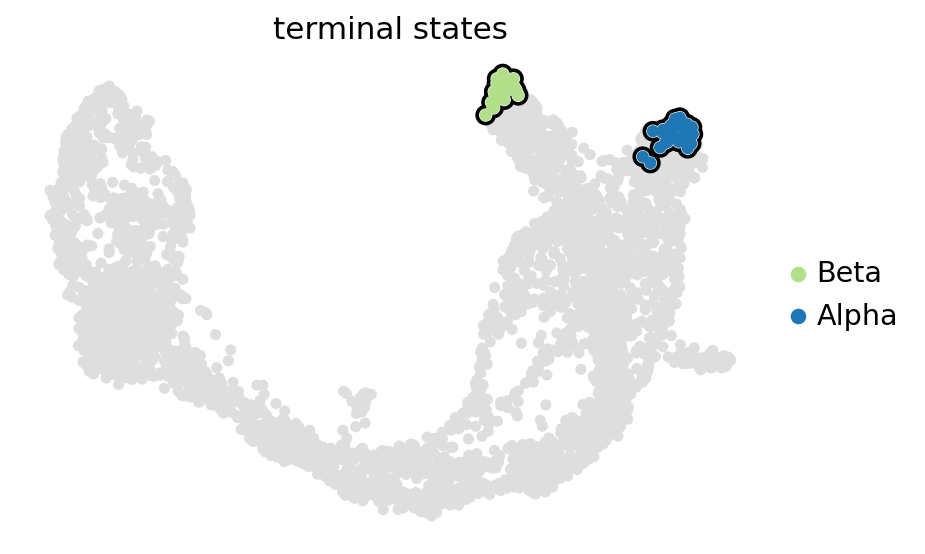
estimator.set_terminal_states(list(set(estimator.macrostates.cat.categories.tolist()).intersection(TERMINAL_STATES)))
GPCCA[kernel=(0.8 * VelocityKernel[n=3696] + 0.2 * ConnectivityKernel[n=3696]), initial_states=None, terminal_states=['Alpha', 'Beta', 'Delta']]
estimator.plot_macrostates(which="terminal", basis="umap", legend_loc="right", s=100)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/scvelo/plotting/scatter.py:656: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
smp = ax.scatter(
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/scvelo/plotting/scatter.py:694: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
ax.scatter(
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/scvelo/plotting/utils.py:1391: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
ax.scatter(x, y, s=bg_size, marker=".", c=bg_color, zorder=zord - 2, **kwargs)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/scvelo/plotting/utils.py:1392: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
ax.scatter(x, y, s=gp_size, marker=".", c=gp_color, zorder=zord - 1, **kwargs)
Randomize regulation#
vae = REGVELOVI.load(DATA_DIR / DATASET / "processed" / "rgv_model", adata)
w = vae.module.v_encoder.fc1.weight.data.detach().clone().cpu().numpy()
# Shuffle genes to randomize weights
gene = adata.var.index.tolist()
random.shuffle(gene)
w = pd.DataFrame(w, index=gene, columns=gene)
w = w.loc[adata.var.index, adata.var.index]
# Convert back to tensor and move to GPU
w = torch.tensor(w.values, device="cuda:0")
INFO File /ictstr01/home/icb/weixu.wang/regulatory_velo/data/pancreatic_endocrine/processed/rgv_model/model.pt
already downloaded
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/lightning/fabric/plugins/environments/slurm.py:204: The `srun` command is available on your system but is not used. HINT: If your intention is to run Lightning on SLURM, prepend your python command with `srun` like so: srun python /home/icb/weixu.wang/miniconda3/envs/regvelo_test/li ...
vae.module.v_encoder.fc1.weight.data = w
adata_random_regulation = adata.copy()
set_output(adata_random_regulation, vae, n_samples=30, batch_size=adata.n_obs)
scv.tl.velocity_graph(adata_random_regulation)
computing velocity graph (using 1/112 cores)
[0]PETSC ERROR: ------------------------------------------------------------------------
[0]PETSC ERROR: Caught signal number 13 Broken Pipe: Likely while reading or writing to a socket
[0]PETSC ERROR: Try option -start_in_debugger or -on_error_attach_debugger
[0]PETSC ERROR: or see https://petsc.org/release/faq/#valgrind and https://petsc.org/release/faq/
[0]PETSC ERROR: configure using --with-debugging=yes, recompile, link, and run
[0]PETSC ERROR: to get more information on the crash.
finished (0:00:09) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
with mplscience.style_context():
fig, ax = plt.subplots(figsize=(4, 3))
scv.pl.velocity_embedding_stream(adata_random_regulation, basis="umap", title="", legend_loc=False, ax=ax)
if SAVE_FIGURES:
fig.savefig(
FIG_DIR / DATASET / "randomize_regulation_vector_field.svg", format="svg", transparent=True, bbox_inches="tight"
)
Predict terminal states#
vk = cr.kernels.VelocityKernel(adata_random_regulation).compute_transition_matrix()
ck = cr.kernels.ConnectivityKernel(adata_random_regulation).compute_transition_matrix()
estimator = cr.estimators.GPCCA(0.8 * vk + 0.2 * ck)
## evaluate the fate prob on original space
estimator.compute_macrostates(n_states=N_STATES, cluster_key="clusters")
[0]PETSC ERROR:
[0]PETSC ERROR:
GPCCA[kernel=(0.8 * VelocityKernel[n=3696] + 0.2 * ConnectivityKernel[n=3696]), initial_states=None, terminal_states=None]
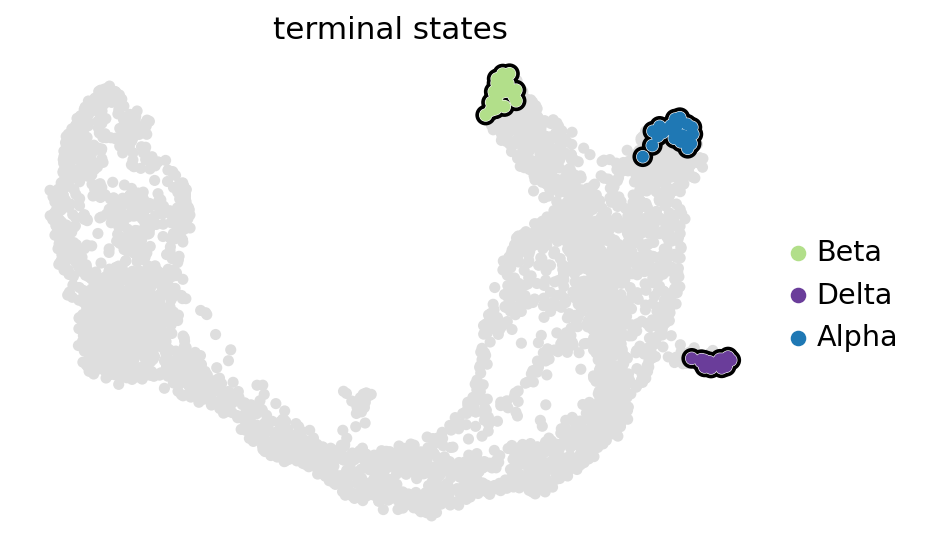
estimator.set_terminal_states(list(set(estimator.macrostates.cat.categories.tolist()).intersection(TERMINAL_STATES)))
GPCCA[kernel=(0.8 * VelocityKernel[n=3696] + 0.2 * ConnectivityKernel[n=3696]), initial_states=None, terminal_states=['Alpha', 'Beta']]
estimator.plot_macrostates(which="terminal", basis="umap", legend_loc="right", s=100)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/scvelo/plotting/scatter.py:656: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
smp = ax.scatter(
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/scvelo/plotting/scatter.py:694: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
ax.scatter(
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/scvelo/plotting/utils.py:1391: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
ax.scatter(x, y, s=bg_size, marker=".", c=bg_color, zorder=zord - 2, **kwargs)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/scvelo/plotting/utils.py:1392: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
ax.scatter(x, y, s=gp_size, marker=".", c=gp_color, zorder=zord - 1, **kwargs)