Performance comparison of inference on dyngen data#
Notebook compares metrics for velocity, latent time and GRN inference across different methods applied to dyngen-generated data.
import numpy as np
import pandas as pd
from scipy.stats import ttest_rel
import matplotlib.pyplot as plt
import mplscience
import seaborn as sns
from rgv_tools import DATA_DIR, FIG_DIR
from rgv_tools.plotting._significance import get_significance
Constants#
DATASET = "dyngen"
SAVE_FIGURES = True
if SAVE_FIGURES:
(FIG_DIR / DATASET).mkdir(parents=True, exist_ok=True)
VELOCITY_METHODS = ["regvelo", "velovi", "scvelo", "velovae_fullvb", "velovae_vae", "tfvelo", "cell2fate", "unitvelo"]
VELOCITY_METHODS_TF = ["regvelo", "tfvelo"]
TIME_METHODS = ["regvelo", "velovi", "scvelo"]
GRN_METHODS = ["regvelo", "tfvelo", "grnboost2", "celloracle", "splicejac", "correlation"]
GRN_METHOD_PALETTE = {
"regvelo": "#0173b2",
"correlation": "#D3D3D3",
"grnboost2": "#D3D3D3", # 708090
"celloracle": "#D3D3D3", # A9A9A9
"tfvelo": "#696969", # 696969
"splicejac": "#D3D3D3",
}
VELO_METHOD_PALETTE = {
"regvelo": "#0173b2",
"velovi": "#de8f05",
"scvelo": "#029e73",
"tfvelo": "#D3D3D3",
"unitvelo": "#D3D3D3",
"velovae_vae": "#D3D3D3",
"velovae_fullvb": "#D3D3D3",
"cell2fate": "#D3D3D3",
}
TIME_METHOD_PALETTE = {
"regvelo": "#0173b2",
"velovi": "#de8f05",
"scvelo": "#029e73",
}
Data loading#
velocity benchmark#
scale_df = []
for scale in range(1, 5):
df_tfvelo = pd.read_parquet(
DATA_DIR / DATASET / f"complexity_{scale}" / "results" / "tfvelo_correlation_all.parquet"
)
velocity_correlation_tf = []
for col in df_tfvelo:
df_tf = df_tfvelo[col]
df_tf = df_tf.reset_index(drop=True)
nan_indices = df_tf[df_tf.isnull()].index
tf_sub = df_tf.drop(nan_indices, axis=0)
n_tf = len(tf_sub)
velocity_correlation_tf.append(np.mean(tf_sub))
correlation_df = []
time_df = []
grn_df = []
for method in VELOCITY_METHODS:
df = pd.read_parquet(DATA_DIR / DATASET / f"complexity_{scale}" / "results" / f"{method}_correlation.parquet")
df.columns = f"{method}_" + df.columns
correlation_df.append(df)
for method in TIME_METHODS:
df = pd.read_parquet(DATA_DIR / DATASET / f"complexity_{scale}" / "results" / f"{method}_correlation.parquet")
df.columns = f"{method}_" + df.columns
time_df.append(df)
for method in GRN_METHODS:
df = pd.read_parquet(DATA_DIR / DATASET / f"complexity_{scale}" / "results" / f"{method}_correlation.parquet")
df.columns = f"{method}_" + df.columns
grn_df.append(df)
correlation_df = pd.concat(correlation_df, axis=1)
time_df = pd.concat(time_df, axis=1)
grn_df = pd.concat(grn_df, axis=1)
if len(velocity_correlation_tf) < correlation_df.shape[0]:
velocity_correlation_tf += [np.nan] * (correlation_df.shape[0] - len(velocity_correlation_tf))
correlation_df["tfvelo_velocity"] = velocity_correlation_tf
df = correlation_df.loc[:, correlation_df.columns.str.contains("velocity")]
df.columns = df.columns.str.removesuffix("_velocity")
df = pd.melt(df, var_name="method", value_name="correlation")
df["correlation"] = (df["correlation"] + 1) / 2
df["scale"] = str(scale)
scale_df.append(df)
df = pd.concat(scale_df, axis=0)
df = df.reset_index(drop=True)
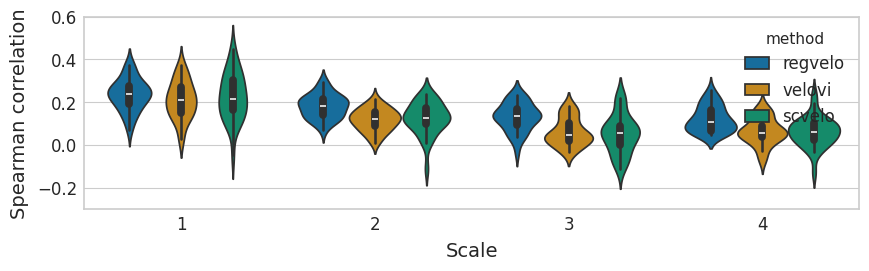
Velocity#
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(14, 2.5))
sns.violinplot(
data=df,
y="correlation",
x="scale",
hue="method",
hue_order=VELOCITY_METHODS,
palette=VELO_METHOD_PALETTE,
ax=ax,
)
ax.set(
xlabel="Pearson correlation",
ylabel="Scale",
yticks=ax.get_yticks(),
)
if SAVE_FIGURES:
fig.savefig(
FIG_DIR / DATASET / "velocity_benchmark.svg",
format="svg",
transparent=True,
bbox_inches="tight",
)
plt.show()
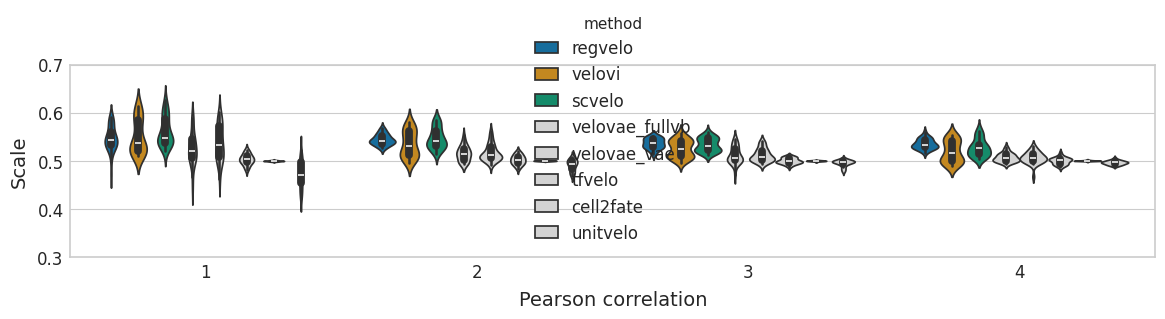
GRN benchmark#
scale_df = []
for scale in range(1, 5):
grn_df = []
for method in GRN_METHODS:
df = pd.read_parquet(f"results/complexity_{scale}/{method}_correlation.parquet")
df.columns = f"{method}_" + df.columns
grn_df.append(df)
grn_df = pd.concat(grn_df, axis=1)
grn_df_sub = grn_df.copy()
grn_df_sub = grn_df_sub.dropna()
grn_df_sub
df = grn_df_sub.loc[:, grn_df.columns.str.contains("grn")]
df.columns = df.columns.str.removesuffix("_grn")
df = pd.melt(df, var_name="method").rename(columns={"value": "correlation"})
df["scale"] = str(scale)
scale_df.append(df)
df = pd.concat(scale_df, axis=0)
df = df.reset_index(drop=True)
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(14, 2.5))
sns.violinplot(
data=df, y="correlation", x="scale", hue="method", hue_order=GRN_METHODS, palette=GRN_METHOD_PALETTE, ax=ax
)
ax.set(
ylabel="AUROC",
xlabel="Scale",
yticks=ax.get_yticks(),
)
if SAVE_FIGURES:
fig.savefig(
FIG_DIR / DATASET / "GRN_benchmark.svg",
format="svg",
transparent=True,
bbox_inches="tight",
)
plt.show()
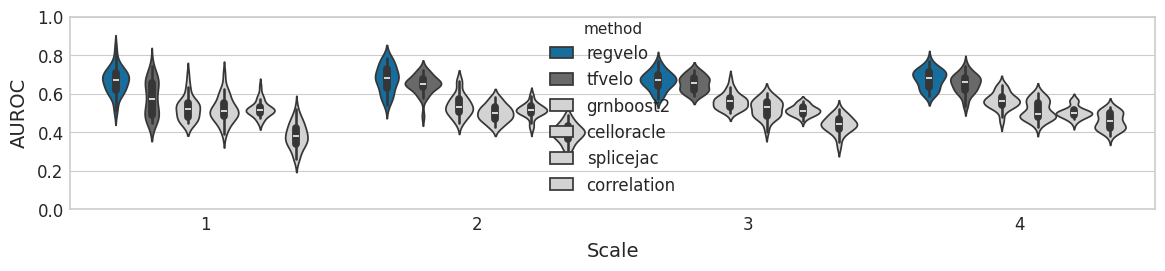
Latent time#
scale_df = []
for scale in range(1, 5):
time_df = []
for method in TIME_METHODS:
df = pd.read_parquet(DATA_DIR / DATASET / f"complexity_{scale}" / "results" / f"{method}_correlation.parquet")
df.columns = f"{method}_" + df.columns
time_df.append(df)
time_df = pd.concat(time_df, axis=1)
ttest_res = ttest_rel(
time_df["regvelo_time"],
time_df["velovi_time"],
alternative="greater",
)
print(ttest_res)
significance = get_significance(pvalue=ttest_res.pvalue)
time_df_sub = time_df.copy()
time_df_sub = time_df_sub.dropna()
time_df_sub
df = time_df_sub.loc[:, time_df.columns.str.contains("time")]
df.columns = df.columns.str.removesuffix("_time")
df = pd.melt(df, var_name="method", value_name="correlation")
df["scale"] = str(scale)
scale_df.append(df)
TtestResult(statistic=2.819809533651689, pvalue=0.004287626705348869, df=29)
TtestResult(statistic=6.817578655205317, pvalue=8.6925544495783e-08, df=29)
TtestResult(statistic=7.716392336845313, pvalue=8.252679776252025e-09, df=29)
TtestResult(statistic=5.498852429313436, pvalue=3.1735361936536186e-06, df=29)
df = pd.concat(scale_df, axis=0)
df = df.reset_index(drop=True)
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(10, 2.5))
sns.violinplot(
data=df, y="correlation", x="scale", hue="method", hue_order=TIME_METHODS, palette=TIME_METHOD_PALETTE, ax=ax
)
ax.set(
ylabel="Spearman correlation",
xlabel="Scale",
yticks=ax.get_yticks(),
)
ax.set_ylim(-0.3, 0.6)
if SAVE_FIGURES:
fig.savefig(
FIG_DIR / DATASET / "time_benchmark.svg",
format="svg",
transparent=True,
bbox_inches="tight",
)
plt.show()