Benchmark transition from hub cells to terminal states#
For simplicity of demonstration, we present the analysis steps for scale-1; the analysis for the other scales is identical.
from itertools import chain
from tqdm import tqdm
import numpy as np
import pandas as pd
from scipy.stats import ttest_ind
import matplotlib.pyplot as plt
import mplscience
import seaborn as sns
import cellrank as cr
import scanpy as sc
import scvelo as scv
from rgv_tools import DATA_DIR, FIG_DIR
from rgv_tools.plotting._significance import add_significance, get_significance
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_csv from `anndata` is deprecated. Import anndata.io.read_csv instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_excel from `anndata` is deprecated. Import anndata.io.read_excel instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_hdf from `anndata` is deprecated. Import anndata.io.read_hdf instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_loom from `anndata` is deprecated. Import anndata.io.read_loom instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_mtx from `anndata` is deprecated. Import anndata.io.read_mtx instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_text from `anndata` is deprecated. Import anndata.io.read_text instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_umi_tools from `anndata` is deprecated. Import anndata.io.read_umi_tools instead.
warnings.warn(msg, FutureWarning)
General setting#
plt.rcParams["svg.fonttype"] = "none"
sns.reset_defaults()
sns.reset_orig()
scv.settings.set_figure_params("scvelo", dpi_save=400, dpi=80, transparent=True, fontsize=14, color_map="viridis")
Constants#
DATASET = "mouse_neural_crest"
SAVE_FIGURES = True
if SAVE_FIGURES:
(FIG_DIR / DATASET).mkdir(parents=True, exist_ok=True)
SAVE_DATA = True
if SAVE_DATA:
(DATA_DIR / DATASET).mkdir(parents=True, exist_ok=True)
FIGURE_FORMATE = "svg"
VELOCITY_METHODS = ["regvelo", "scvelo", "velovi"]
TERMINAL_STATE = ["Melanocytes", "enFib", "SC", "Sensory", "ChC", "SatGlia", "Gut_glia", "Gut_neuron", "Symp", "BCC"]
STATE_TRANSITIONS_RAW = [
("hub", "Sensory"),
("hub", "SatGlia"),
("hub", "SC"),
("hub", "Gut_glia"),
("hub", "Gut_neuron"),
("hub", "ChC"),
]
scale_level = 2
Data loading#
vks = {}
for method in VELOCITY_METHODS:
adata = sc.read_h5ad(DATA_DIR / DATASET / "processed" / f"adata_run_stage_{scale_level}_{method}_all_regulons.h5ad")
adata.obs["Hub"] = (~adata.obs["conflict"].astype(bool)) & (adata.obs["assignments"] == "none")
adata.obs["assignments"] = adata.obs["assignments"].astype(str)
adata.obs["assignments"][adata.obs["Hub"]] = "hub"
print(adata)
## construct graph
vk = cr.kernels.VelocityKernel(adata).compute_transition_matrix()
vks[method] = vk
AnnData object with n_obs × n_vars = 3926 × 1122
obs: 'plates', 'devtime', 'location', 'n_genes_by_counts', 'total_counts', 'total_counts_ERCC', 'pct_counts_ERCC', 'doublet_scores', 'leiden', 'CytoTRACE', 'Gut_neuron', 'Sensory', 'Symp', 'enFib', 'ChC', 'Gut_glia', 'NCC', 'Mesenchyme', 'Melanocytes', 'SatGlia', 'SC', 'BCC', 'conflict', 'assignments', 'batch', 'initial_size_unspliced', 'initial_size_spliced', 'initial_size', 'n_counts', 'Hub'
var: 'ERCC', 'n_cells_by_counts', 'mean_counts', 'pct_dropout_by_counts', 'total_counts', 'n_cells', 'm', 'v', 'n_obs', 'res', 'lp', 'lpa', 'qv', 'highly_variable', 'Accession', 'Chromosome', 'End', 'Start', 'Strand', 'TF', 'means', 'dispersions', 'dispersions_norm', 'velocity_genes', 'fit_beta', 'fit_gamma', 'fit_scaling'
uns: '_scvi_manager_uuid', '_scvi_uuid', 'assignments_colors', 'devtime_colors', 'hvg', 'leiden', 'leiden_colors', 'leiden_sizes', 'location_colors', 'log1p', 'neighbors', 'network', 'paga', 'regulators', 'skeleton', 'targets', 'umap'
obsm: 'X_diff', 'X_pca', 'X_umap'
layers: 'GEX', 'Ms', 'Mu', 'ambiguous', 'fit_t', 'latent_time_velovi', 'matrix', 'palantir_imp', 'scaled', 'spanning', 'spliced', 'unspliced', 'velocity'
obsp: 'connectivities', 'distances'
AnnData object with n_obs × n_vars = 3926 × 1122
obs: 'plates', 'devtime', 'location', 'n_genes_by_counts', 'total_counts', 'total_counts_ERCC', 'pct_counts_ERCC', 'doublet_scores', 'leiden', 'CytoTRACE', 'Gut_neuron', 'Sensory', 'Symp', 'enFib', 'ChC', 'Gut_glia', 'NCC', 'Mesenchyme', 'Melanocytes', 'SatGlia', 'SC', 'BCC', 'conflict', 'assignments', 'batch', 'initial_size_unspliced', 'initial_size_spliced', 'initial_size', 'n_counts', 'Hub'
var: 'ERCC', 'n_cells_by_counts', 'mean_counts', 'pct_dropout_by_counts', 'total_counts', 'n_cells', 'm', 'v', 'n_obs', 'res', 'lp', 'lpa', 'qv', 'highly_variable', 'Accession', 'Chromosome', 'End', 'Start', 'Strand', 'TF', 'means', 'dispersions', 'dispersions_norm', 'velocity_genes', 'fit_alpha', 'fit_beta', 'fit_gamma', 'fit_t_', 'fit_scaling', 'fit_std_u', 'fit_std_s', 'fit_likelihood', 'fit_u0', 'fit_s0', 'fit_pval_steady', 'fit_steady_u', 'fit_steady_s', 'fit_variance', 'fit_alignment_scaling'
uns: 'assignments_colors', 'devtime_colors', 'hvg', 'leiden', 'leiden_colors', 'leiden_sizes', 'location_colors', 'log1p', 'neighbors', 'network', 'paga', 'recover_dynamics', 'regulators', 'skeleton', 'targets', 'umap', 'velocity_params'
obsm: 'X_diff', 'X_pca', 'X_umap'
varm: 'loss'
layers: 'GEX', 'Ms', 'Mu', 'ambiguous', 'fit_t', 'fit_tau', 'fit_tau_', 'matrix', 'palantir_imp', 'scaled', 'spanning', 'spliced', 'unspliced', 'velocity', 'velocity_u'
obsp: 'connectivities', 'distances'
AnnData object with n_obs × n_vars = 3926 × 1122
obs: 'plates', 'devtime', 'location', 'n_genes_by_counts', 'total_counts', 'total_counts_ERCC', 'pct_counts_ERCC', 'doublet_scores', 'leiden', 'CytoTRACE', 'Gut_neuron', 'Sensory', 'Symp', 'enFib', 'ChC', 'Gut_glia', 'NCC', 'Mesenchyme', 'Melanocytes', 'SatGlia', 'SC', 'BCC', 'conflict', 'assignments', 'batch', 'initial_size_unspliced', 'initial_size_spliced', 'initial_size', 'n_counts', 'Hub'
var: 'ERCC', 'n_cells_by_counts', 'mean_counts', 'pct_dropout_by_counts', 'total_counts', 'n_cells', 'm', 'v', 'n_obs', 'res', 'lp', 'lpa', 'qv', 'highly_variable', 'Accession', 'Chromosome', 'End', 'Start', 'Strand', 'TF', 'means', 'dispersions', 'dispersions_norm', 'velocity_genes', 'fit_alpha', 'fit_beta', 'fit_gamma', 'fit_scaling'
uns: '_scvi_manager_uuid', '_scvi_uuid', 'assignments_colors', 'devtime_colors', 'hvg', 'leiden', 'leiden_colors', 'leiden_sizes', 'location_colors', 'log1p', 'neighbors', 'network', 'paga', 'regulators', 'skeleton', 'targets', 'umap'
obsm: 'X_diff', 'X_pca', 'X_umap'
layers: 'GEX', 'Ms', 'Mu', 'ambiguous', 'fit_t', 'latent_time_velovi', 'matrix', 'palantir_imp', 'scaled', 'spanning', 'spliced', 'unspliced', 'velocity'
obsp: 'connectivities', 'distances'
Filter out the transition#
STATE_TRANSITIONS = []
for i in range(len(STATE_TRANSITIONS_RAW)):
if STATE_TRANSITIONS_RAW[i][1] in np.unique(adata.obs["assignments"]):
STATE_TRANSITIONS.append(STATE_TRANSITIONS_RAW[i])
STATE_TRANSITIONS_RAW = [f"{a} - {b}" for a, b in STATE_TRANSITIONS_RAW]
STATE_TRANSITIONS
[('hub', 'Sensory'), ('hub', 'SatGlia')]
Measuring transport from hub to the terminal cell states#
STATE_TRANSITIONS
[('hub', 'Sensory'), ('hub', 'SatGlia')]
cluster_key = "assignments"
rep = "X_pca"
score_df_rgv_vs_scv = []
score_df_rgv_vs_vi = []
cbc_rgv_f = []
cbc_scv_f = []
cbc_vi_f = []
for source, target in tqdm(STATE_TRANSITIONS):
cbc_rgv = vks["regvelo"].cbc(source=source, target=target, cluster_key=cluster_key, rep=rep)
cbc_scv = vks["scvelo"].cbc(source=source, target=target, cluster_key=cluster_key, rep=rep)
cbc_vi = vks["velovi"].cbc(source=source, target=target, cluster_key=cluster_key, rep=rep)
score_df_rgv_vs_scv.append(
pd.DataFrame(
{
"State transition": [f"{source} - {target}"] * len(cbc_rgv),
"Log ratio": np.log((cbc_rgv + 1) / (cbc_scv + 1)),
}
)
)
score_df_rgv_vs_vi.append(
pd.DataFrame(
{
"State transition": [f"{source} - {target}"] * len(cbc_rgv),
"Log ratio": np.log((cbc_rgv + 1) / (cbc_vi + 1)),
}
)
)
cbc_rgv_f.append(cbc_rgv)
cbc_scv_f.append(cbc_scv)
cbc_vi_f.append(cbc_vi)
score_df_rgv_vs_scv_forward = pd.concat(score_df_rgv_vs_scv)
score_df_rgv_vs_vi_forward = pd.concat(score_df_rgv_vs_vi)
100%|██████████| 2/2 [00:00<00:00, 4.71it/s]
Comparing overall transition#
dfs = []
g_df = pd.DataFrame({"CBC_forward": list(chain(*cbc_rgv_f))})
g_df["Method"] = "RegVelo"
dfs.append(g_df)
g_df = pd.DataFrame({"CBC_forward": list(chain(*cbc_vi_f))})
g_df["Method"] = "veloVI"
dfs.append(g_df)
g_df = pd.DataFrame({"CBC_forward": list(chain(*cbc_scv_f))})
g_df["Method"] = "scVelo"
dfs.append(g_df)
velo_df = pd.concat(dfs, axis=0)
velo_df["Method"] = velo_df["Method"].astype("category")
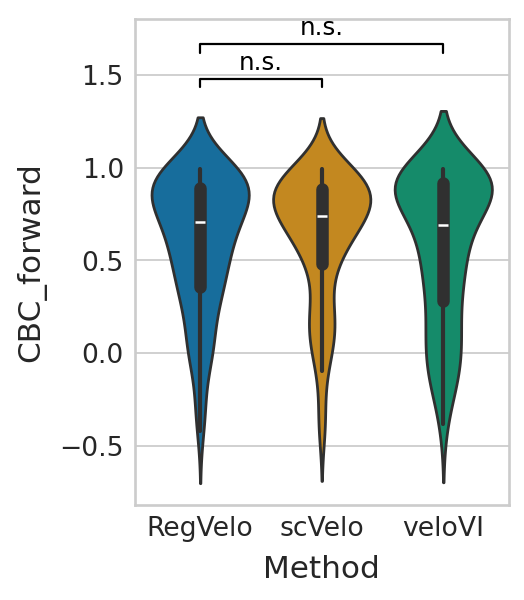
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(3, 4))
sns.violinplot(data=velo_df, x="Method", y="CBC_forward", palette="colorblind", ax=ax)
ttest_res = ttest_ind(list(chain(*cbc_rgv_f)), list(chain(*cbc_vi_f)), equal_var=False, alternative="greater")
significance = get_significance(ttest_res.pvalue)
add_significance(
ax=ax,
left=0,
right=1,
significance=significance,
lw=1,
bracket_level=1.05,
c="k",
level=0,
)
ttest_res = ttest_ind(list(chain(*cbc_rgv_f)), list(chain(*cbc_scv_f)), equal_var=False, alternative="greater")
significance = get_significance(ttest_res.pvalue)
add_significance(
ax=ax,
left=0,
right=2,
significance=significance,
lw=1,
bracket_level=1.05,
c="k",
level=0,
)
y_min, y_max = ax.get_ylim()
ax.set_ylim([y_min, y_max + 0.02])
plt.show()
if SAVE_DATA:
velo_df.to_csv(DATA_DIR / DATASET / "velo_df_scale_1.csv")
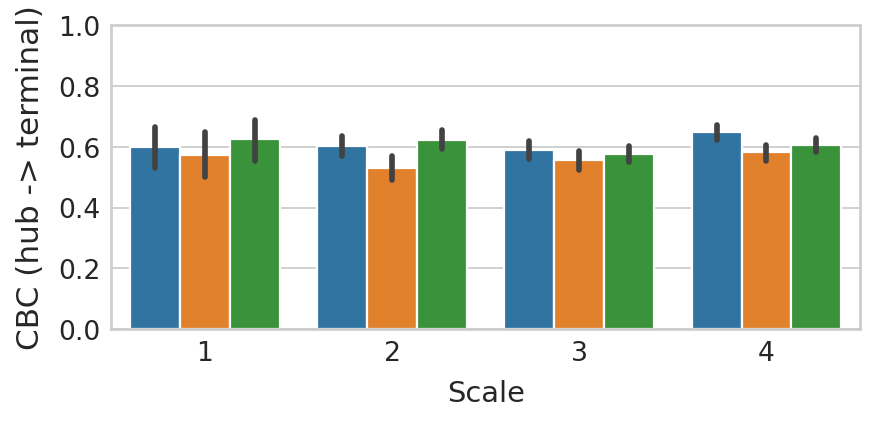
Visualize all transition results#
df_all = []
for i in range(1, 5):
df = pd.read_csv(DATA_DIR / DATASET / f"velo_df_scale_{i}.csv", index_col=0)
df["scale"] = str(i)
df_all.append(df)
df = pd.concat(df_all, axis=0)
df
| CBC_forward | Method | scale | |
|---|---|---|---|
| 0 | 0.981314 | RegVelo | 1 |
| 1 | 0.476706 | RegVelo | 1 |
| 2 | 0.753209 | RegVelo | 1 |
| 3 | 0.548564 | RegVelo | 1 |
| 4 | 0.612676 | RegVelo | 1 |
| ... | ... | ... | ... |
| 609 | 0.491018 | scVelo | 4 |
| 610 | 0.174424 | scVelo | 4 |
| 611 | 0.297567 | scVelo | 4 |
| 612 | -0.316798 | scVelo | 4 |
| 613 | 0.165285 | scVelo | 4 |
4278 rows × 3 columns
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(6, 2.5))
# Plot the barplot without error bars
sns.barplot(data=df, x="scale", y="CBC_forward", hue="Method", ax=ax)
# Add jittered dots
# sns.stripplot(data=df, y="scale", x="AUROC", hue="method", dodge=True, color="black", ax=ax, jitter=True)
# Remove the duplicate legend
handles, labels = ax.get_legend_handles_labels()
ax.legend(handles[3:6], labels[3:6], bbox_to_anchor=(0.5, -0.1), loc="upper center", ncol=2)
# Customize labels and other settings
ax.set(xlabel="", ylabel="CBC (hub -> terminal)")
ax.set_xlabel(xlabel="Scale", fontsize=13)
ax.set_ylim(0, 1)
if SAVE_FIGURES:
plt.savefig(FIG_DIR / DATASET / "CBC_benchmark.svg", format="svg", transparent=True, bbox_inches="tight")
plt.show()