Benchmark perturbation prediction#
notebook for benchmarking perturbation prediction
Library imports#
import numpy as np
import pandas as pd
import scipy
import matplotlib.pyplot as plt
import mplscience
import seaborn as sns
import scvi
from rgv_tools import DATA_DIR, FIG_DIR
from rgv_tools.core import METHOD_PALETTE_PERTURBATION
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_csv from `anndata` is deprecated. Import anndata.io.read_csv instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_excel from `anndata` is deprecated. Import anndata.io.read_excel instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_hdf from `anndata` is deprecated. Import anndata.io.read_hdf instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_loom from `anndata` is deprecated. Import anndata.io.read_loom instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_mtx from `anndata` is deprecated. Import anndata.io.read_mtx instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_text from `anndata` is deprecated. Import anndata.io.read_text instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_umi_tools from `anndata` is deprecated. Import anndata.io.read_umi_tools instead.
warnings.warn(msg, FutureWarning)
General setting#
scvi.settings.seed = 0
[rank: 0] Seed set to 0
DATASET = "zebrafish"
SAVE_FIGURES = True
if SAVE_FIGURES:
(FIG_DIR / DATASET / "results").mkdir(parents=True, exist_ok=True)
Constant#
single_ko = ["rarga", "rxraa", "nr2f5", "fli1a", "tfec", "elk3", "mitfa", "ets1", "nr2f2", "elf1", "ebf3a"]
multiple_ko = ["fli1a_elk3", "tfec_mitfa_bhlhe40", "mitfa_tfec", "mitfa_tfec_tfeb"]
terminal_states = ["mNC_arch2", "mNC_head_mesenchymal", "mNC_hox34", "Pigment"]
METHODS = ["RegVelo", "Dynamo (KO)", "Dynamo (perturbation)", "celloracle"]
GENE_MARKER = {
"rarga": "",
"rxraa": "o",
"nr2f5": "o",
"fli1a": "o",
"tfec": "o",
"elk3": "",
"mitfa": "o",
"ets1": "X",
"nr2f2": (4, 0, 45),
"elf1": "P",
"ebf3a": (4, 0, 0),
"fli1a_elk3": (4, 1, 0),
"tfec_mitfa_bhlhe40": "^",
"mitfa_tfec": (4, 1, 45),
"mitfa_tfec_tfeb": "v",
}
Data loading#
Perturbation predicton#
perturbation = pd.read_csv(DATA_DIR / DATASET / "results" / "MELD_median_score.csv", index_col=0)
TERMINAL_STATES_KO = perturbation.columns.tolist()
TERMINAL_STATES_PERTURB = perturbation.columns.tolist()
TERMINAL_STATES_PERTURB[3] = "Pigment"
TERMINAL_STATES_KO[3] = "Pigment_gch2"
Single knock-out panel prediction results#
coef_perturb_co = pd.read_csv(DATA_DIR / DATASET / "results" / "celloracle_perturb_single.csv", index_col=0)
coef_perturb_dyn = pd.read_csv(DATA_DIR / DATASET / "results" / "dynamo_perturb_single.csv", index_col=0)
coef_KO_dyn = pd.read_csv(DATA_DIR / DATASET / "results" / "dynamo_KO_single.csv", index_col=0)
coef_perturb_co = coef_perturb_co.loc[single_ko, TERMINAL_STATES_PERTURB]
coef_perturb_dyn = coef_perturb_dyn.loc[single_ko, TERMINAL_STATES_PERTURB]
coef_KO_dyn = coef_KO_dyn.loc[single_ko, TERMINAL_STATES_KO]
single_matrix = perturbation.loc[single_ko, :]
score_s = []
method = []
score_s.append(scipy.stats.spearmanr(np.array(single_matrix).reshape(-1), np.array(1 - coef_perturb_co).reshape(-1))[0])
method.append("celloracle")
score_s.append(scipy.stats.spearmanr(np.array(single_matrix).reshape(-1), np.array(coef_perturb_dyn).reshape(-1))[0])
method.append("Dynamo (perturbation)")
score_s.append(scipy.stats.spearmanr(np.array(single_matrix).reshape(-1), np.array(coef_KO_dyn).reshape(-1))[0])
method.append("Dynamo (KO)")
for nrun in range(3):
coef_perturb_rgv = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_single.csv", index_col=0
)
coef_perturb_rgv = coef_perturb_rgv.loc[single_ko, TERMINAL_STATES_KO]
score_s.append(
scipy.stats.spearmanr(np.array(single_matrix).reshape(-1), np.array(coef_perturb_rgv).reshape(-1))[0]
)
method.append("RegVelo")
Add multiple knock-out panel prediction results#
coef_perturb_co = pd.read_csv(DATA_DIR / DATASET / "results" / "celloracle_perturb_single.csv", index_col=0)
coef_perturb_dyn = pd.read_csv(DATA_DIR / DATASET / "results" / "dynamo_perturb_single.csv", index_col=0)
coef_KO_dyn = pd.read_csv(DATA_DIR / DATASET / "results" / "dynamo_KO_single.csv", index_col=0)
coef_perturb_co2 = pd.read_csv(DATA_DIR / DATASET / "results" / "celloracle_perturb_multiple.csv", index_col=0)
coef_perturb_dyn2 = pd.read_csv(DATA_DIR / DATASET / "results" / "dynamo_perturb_multiple.csv", index_col=0)
coef_KO_dyn2 = pd.read_csv(DATA_DIR / DATASET / "results" / "dynamo_KO_multiple.csv", index_col=0)
coef_perturb_co = pd.concat([coef_perturb_co, coef_perturb_co2])
coef_perturb_dyn = pd.concat([coef_perturb_dyn, coef_perturb_dyn2])
coef_KO_dyn = pd.concat([coef_KO_dyn, coef_KO_dyn2])
coef_perturb_co = coef_perturb_co.loc[single_ko + multiple_ko, TERMINAL_STATES_PERTURB]
coef_perturb_dyn = coef_perturb_dyn.loc[single_ko + multiple_ko, TERMINAL_STATES_PERTURB]
coef_KO_dyn = coef_KO_dyn.loc[single_ko + multiple_ko, TERMINAL_STATES_KO]
single_matrix = perturbation.loc[single_ko + multiple_ko, :]
score_m = []
method = []
score_m.append(scipy.stats.spearmanr(np.array(single_matrix).reshape(-1), np.array(1 - coef_perturb_co).reshape(-1))[0])
method.append("celloracle")
score_m.append(scipy.stats.spearmanr(np.array(single_matrix).reshape(-1), np.array(coef_perturb_dyn).reshape(-1))[0])
method.append("Dynamo (perturbation)")
score_m.append(scipy.stats.spearmanr(np.array(single_matrix).reshape(-1), np.array(coef_KO_dyn).reshape(-1))[0])
method.append("Dynamo (KO)")
for nrun in range(3):
coef_perturb_rgv = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_single.csv", index_col=0
)
coef_perturb_rgv2 = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_multiple.csv", index_col=0
)
coef_perturb_rgv = pd.concat([coef_perturb_rgv, coef_perturb_rgv2])
coef_perturb_rgv = coef_perturb_rgv.loc[single_ko + multiple_ko, TERMINAL_STATES_KO]
score_m.append(
scipy.stats.spearmanr(np.array(single_matrix).reshape(-1), np.array(coef_perturb_rgv).reshape(-1))[0]
)
method.append("RegVelo")
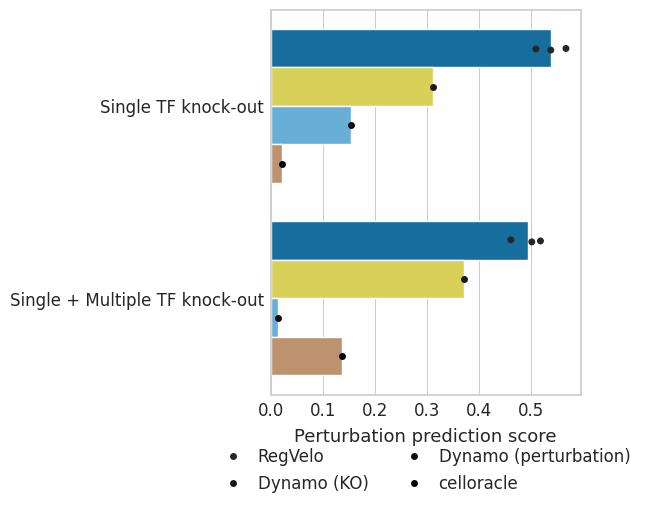
Plot comparsion#
dat = pd.DataFrame({"Spearman_correlation": score_s, "method": method})
dat2 = pd.DataFrame({"Spearman_correlation": score_m, "method": method})
dat["Experimental class"] = "Single TF knock-out"
dat2["Experimental class"] = "Single + Multiple TF knock-out"
df = pd.concat([dat, dat2], axis=0)
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(4, 5))
# Plot the barplot without error bars
sns.barplot(
data=df,
y="Experimental class",
x="Spearman_correlation",
hue="method",
hue_order=METHODS,
palette=METHOD_PALETTE_PERTURBATION,
ax=ax,
ci=None,
)
# Add jittered dots
sns.stripplot(
data=df,
y="Experimental class",
x="Spearman_correlation",
hue="method",
hue_order=METHODS,
dodge=True,
color="black",
ax=ax,
jitter=True,
)
# Remove the duplicate legend
handles, labels = ax.get_legend_handles_labels()
ax.legend(
handles[4 : (4 + len(METHOD_PALETTE_PERTURBATION))],
labels[4 : (4 + len(METHOD_PALETTE_PERTURBATION))],
bbox_to_anchor=(0.5, -0.1),
loc="upper center",
ncol=2,
)
# Customize labels and other settings
ax.set(ylabel="", xlabel="Perturbation prediction score")
ax.set_xlabel(xlabel="Perturbation prediction score", fontsize=13)
if SAVE_FIGURES:
plt.savefig(
FIG_DIR / DATASET / "results" / "barplot_joint_knockout_correlation.svg",
format="svg",
transparent=True,
bbox_inches="tight",
)
plt.show()
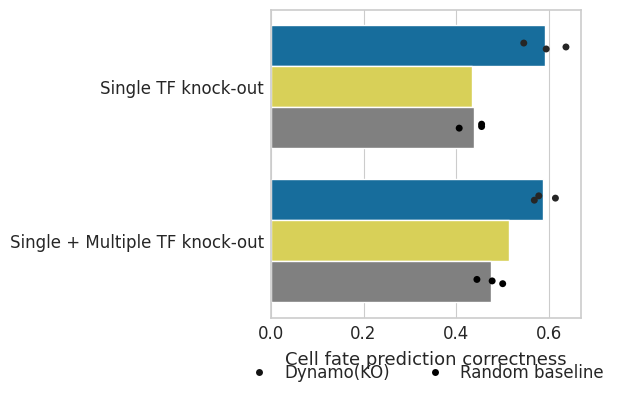
Cell fate prediction precision#
Single knock-out panel prediction results#
single_matrix = perturbation.loc[single_ko, :]
Calculate pvalue#
pval = pd.read_csv(DATA_DIR / DATASET / "results" / "pval_mat.csv", index_col=0)
pval
| mNC_arch2 | mNC_head_mesenchymal | mNC_hox34 | Pigment_gch2_high | |
|---|---|---|---|---|
| fli1a_erf_erfl3 | 0.952 | 0.442 | 0.000 | 1.000 |
| erf_erfl3 | 0.702 | 1.000 | 0.000 | 1.000 |
| mitfa | 0.988 | 1.000 | 0.538 | 1.000 |
| rarga | 0.992 | 0.648 | 0.768 | 0.000 |
| nr2f5 | 0.998 | 1.000 | 0.990 | 1.000 |
| tfec | 0.836 | 0.052 | 0.916 | 1.000 |
| rxraa | 1.000 | 0.412 | 0.724 | 1.000 |
| fli1a_elk3 | 1.000 | 1.000 | 0.024 | 1.000 |
| tfec_mitfa_bhlhe40 | 0.002 | 0.362 | 0.006 | 1.000 |
| elk3 | 0.978 | 0.716 | 0.002 | 1.000 |
| mitfa_tfec | 0.000 | 1.000 | 0.016 | 1.000 |
| mitfa_tfec_tfeb | 1.000 | 0.982 | 0.330 | 1.000 |
| fli1a | 1.000 | 0.000 | 0.000 | 1.000 |
| ebf3a | 0.960 | 0.110 | 0.974 | 1.000 |
| elf1 | 0.466 | 0.004 | 0.782 | 1.000 |
| nr2f2 | 0.762 | 0.998 | 0.856 | 0.012 |
| ets1 | 1.000 | 0.998 | 0.762 | 1.000 |
pval = pval.loc[single_matrix.index.tolist(), single_matrix.columns.tolist()]
mask_pos = (pval <= 0.05).astype(int)
mask_neg = (pval >= 0.95).astype(int)
single_matrix = mask_neg * (-1) + mask_pos
fate_s = []
method2 = []
coef_KO_dyn = pd.read_csv(DATA_DIR / DATASET / "results" / "dynamo_KO_single.csv", index_col=0)
pval_KO_dyn = pd.read_csv(DATA_DIR / DATASET / "results" / "dynamo_KO_single_pval.csv", index_col=0)
coef_KO_dyn = coef_KO_dyn.loc[single_ko, TERMINAL_STATES_KO]
pval_KO_dyn = pval_KO_dyn.loc[single_ko, TERMINAL_STATES_KO]
pred_m = coef_KO_dyn - 0.5
pred_m_filter = pred_m[pval_KO_dyn < 0.05]
pred_m_filter[np.isnan(pred_m_filter)] = 0
pred_m_filter.columns = single_matrix.columns
score = np.sum(np.sign(single_matrix) * np.sign(pred_m_filter) == 1).sum() / np.sum(pred_m_filter != 0).sum()
fate_s.append(score)
method2.append("Dynamo (KO)")
for nrun in range(3):
coef_perturb_rgv = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_single.csv", index_col=0
)
pval_perturb_rgv = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_single_pval.csv", index_col=0
)
coef_perturb_rgv = coef_perturb_rgv.loc[single_ko, TERMINAL_STATES_KO]
pval_perturb_rgv = pval_perturb_rgv.loc[single_ko, TERMINAL_STATES_KO]
pred_m = coef_perturb_rgv
pred_m_filter = pred_m[pval_perturb_rgv < 0.05]
pred_m_filter[np.isnan(pred_m_filter)] = 0
pred_m_filter.columns = single_matrix.columns
score = np.sum(np.sign(single_matrix) * np.sign(pred_m_filter) == 1).sum() / np.sum(pred_m_filter != 0).sum()
fate_s.append(score)
method2.append("RegVelo")
for nrun in range(3):
coef_perturb_rgv = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_single.csv", index_col=0
)
pval_perturb_rgv = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_single_pval.csv", index_col=0
)
coef_perturb_rgv = coef_perturb_rgv.loc[single_ko, TERMINAL_STATES_KO]
pval_perturb_rgv = pval_perturb_rgv.loc[single_ko, TERMINAL_STATES_KO]
coef_perturb_rgv = coef_perturb_rgv.sample(frac=1).reset_index(drop=True)
pval_perturb_rgv = pval_perturb_rgv.sample(frac=1).reset_index(drop=True)
pred_m = coef_perturb_rgv
pred_m_filter = pred_m[pval_perturb_rgv < 0.05]
pred_m_filter[np.isnan(pred_m_filter)] = 0
pred_m_filter.columns = single_matrix.columns
pred_m_filter.index = single_matrix.index
# shuffled = np.random.permutation(pred_m_filter)
score = np.sum(np.sign(single_matrix) * np.sign(pred_m_filter) == 1).sum() / np.sum(pred_m_filter != 0).sum()
fate_s.append(score)
method2.append("Random baseline")
Multiple knock-out panel prediction results#
single_matrix = perturbation.loc[single_ko + multiple_ko, :]
pval = pd.read_csv(DATA_DIR / DATASET / "results" / "pval_mat.csv", index_col=0)
pval = pval.loc[single_matrix.index.tolist(), single_matrix.columns.tolist()]
mask_pos = (pval <= 0.05).astype(int)
mask_neg = (pval >= 0.95).astype(int)
single_matrix = mask_neg * (-1) + mask_pos
coef_KO_dyn = pd.read_csv(DATA_DIR / DATASET / "results" / "dynamo_KO_single.csv", index_col=0)
coef_KO_dyn2 = pd.read_csv(DATA_DIR / DATASET / "results" / "dynamo_KO_multiple.csv", index_col=0)
coef_KO_dyn = pd.concat([coef_KO_dyn, coef_KO_dyn2])
coef_KO_dyn = coef_KO_dyn.loc[single_ko + multiple_ko, TERMINAL_STATES_KO]
fate_m = []
method2 = []
pval_KO_dyn = pd.read_csv(DATA_DIR / DATASET / "results" / "dynamo_KO_single_pval.csv", index_col=0)
pval_KO_dyn2 = pd.read_csv(DATA_DIR / DATASET / "results" / "dynamo_KO_multiple_pval.csv", index_col=0)
pval_KO_dyn = pd.concat([pval_KO_dyn, pval_KO_dyn2])
pval_KO_dyn = pval_KO_dyn.loc[single_ko + multiple_ko, TERMINAL_STATES_KO]
pred_m = coef_KO_dyn - 0.5
pred_m_filter = pred_m[pval_KO_dyn < 0.05]
pred_m_filter[np.isnan(pred_m_filter)] = 0
pred_m_filter.columns = single_matrix.columns
score = np.sum(np.sign(single_matrix) * np.sign(pred_m_filter) == 1).sum() / np.sum(pred_m_filter != 0).sum()
fate_m.append(score)
method2.append("Dynamo (KO)")
for nrun in range(3):
coef_perturb_rgv = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_single.csv", index_col=0
)
coef_perturb_rgv2 = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_multiple.csv", index_col=0
)
coef_perturb_rgv = pd.concat([coef_perturb_rgv, coef_perturb_rgv2])
pval_perturb_rgv = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_single_pval.csv", index_col=0
)
pval_perturb_rgv2 = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_multiple_pval.csv", index_col=0
)
pval_perturb_rgv = pd.concat([pval_perturb_rgv, pval_perturb_rgv2])
coef_perturb_rgv = coef_perturb_rgv.loc[single_ko + multiple_ko, TERMINAL_STATES_KO]
pval_perturb_rgv = pval_perturb_rgv.loc[single_ko + multiple_ko, TERMINAL_STATES_KO]
pred_m = coef_perturb_rgv
pred_m_filter = pred_m[pval_perturb_rgv < 0.05]
pred_m_filter[np.isnan(pred_m_filter)] = 0
pred_m_filter.columns = single_matrix.columns
score = np.sum(np.sign(single_matrix) * np.sign(pred_m_filter) == 1).sum() / np.sum(pred_m_filter != 0).sum()
fate_m.append(score)
method2.append("RegVelo")
for nrun in range(3):
coef_perturb_rgv = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_single.csv", index_col=0
)
coef_perturb_rgv2 = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_multiple.csv", index_col=0
)
coef_perturb_rgv = pd.concat([coef_perturb_rgv, coef_perturb_rgv2])
pval_perturb_rgv = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_single_pval.csv", index_col=0
)
pval_perturb_rgv2 = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_multiple_pval.csv", index_col=0
)
pval_perturb_rgv = pd.concat([pval_perturb_rgv, pval_perturb_rgv2])
coef_perturb_rgv = coef_perturb_rgv.loc[single_ko + multiple_ko, TERMINAL_STATES_KO]
pval_perturb_rgv = pval_perturb_rgv.loc[single_ko + multiple_ko, TERMINAL_STATES_KO]
coef_perturb_rgv = coef_perturb_rgv.sample(frac=1).reset_index(drop=True)
pval_perturb_rgv = pval_perturb_rgv.sample(frac=1).reset_index(drop=True)
pred_m = coef_perturb_rgv
pred_m_filter = pred_m[pval_perturb_rgv < 0.05]
pred_m_filter[np.isnan(pred_m_filter)] = 0
pred_m_filter.columns = single_matrix.columns
pred_m_filter.index = single_matrix.index
# shuffled = np.random.permutation(pred_m_filter)
score = np.sum(np.sign(single_matrix) * np.sign(pred_m_filter) == 1).sum() / np.sum(pred_m_filter != 0).sum()
fate_m.append(score)
method2.append("Random baseline")
Visualize prediction results#
dat = pd.DataFrame({"cell fate prediction correctness": fate_s, "method": method2})
dat2 = pd.DataFrame({"cell fate prediction correctness": fate_m, "method": method2})
dat["Experimental class"] = "Single TF knock-out"
dat2["Experimental class"] = "Single + Multiple TF knock-out"
df = pd.concat([dat, dat2], axis=0)
METHOD_PALETTE_PERTURBATION["Random baseline"] = "#808080"
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(4, 4))
# Plot the barplot without error bars
sns.barplot(
data=df,
y="Experimental class",
x="cell fate prediction correctness",
hue="method",
hue_order=["RegVelo", "Dynamo (KO)", "Random baseline"],
palette=METHOD_PALETTE_PERTURBATION,
ax=ax,
ci=None,
)
# Add jittered dots
sns.stripplot(
data=df,
y="Experimental class",
x="cell fate prediction correctness",
hue="method",
hue_order=["RegVelo", "Dynamo(KO)", "Random baseline"],
dodge=True,
color="black",
ax=ax,
jitter=True,
)
# Remove the duplicate legend
handles, labels = ax.get_legend_handles_labels()
ax.legend(
handles[4 : (4 + len(METHOD_PALETTE_PERTURBATION))],
labels[4 : (4 + len(METHOD_PALETTE_PERTURBATION))],
bbox_to_anchor=(0.5, -0.1),
loc="upper center",
ncol=2,
)
# Customize labels and other settings
ax.set(ylabel="", xlabel="Cell fate prediction precision")
ax.set_xlabel(xlabel="Cell fate prediction correctness", fontsize=13)
# ax.set_xlim(0.45,)
if SAVE_FIGURES:
plt.savefig(
DATA_DIR / DATASET / "results" / "cell_fate_precision.svg",
format="svg",
transparent=True,
bbox_inches="tight",
)
plt.show()
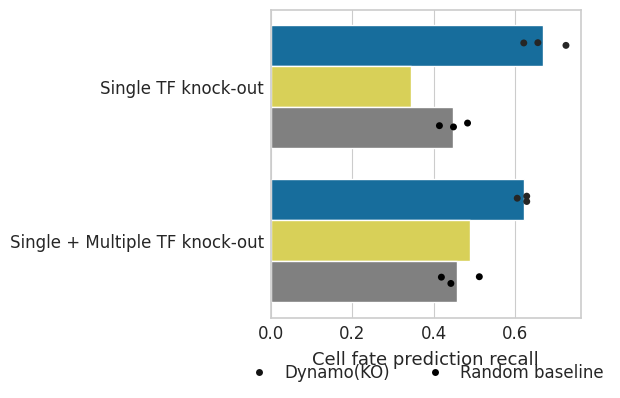
Cell fate prediction Recall#
Single knock-out panel prediction results#
single_matrix = perturbation.loc[single_ko, :]
Calculate pvalue#
pval = pd.read_csv(DATA_DIR / DATASET / "results" / "pval_mat.csv", index_col=0)
pval = pval.loc[single_matrix.index.tolist(), single_matrix.columns.tolist()]
mask_pos = (pval <= 0.05).astype(int)
mask_neg = (pval >= 0.95).astype(int)
single_matrix = mask_neg * (-1) + mask_pos
fate_s = []
method2 = []
coef_KO_dyn = pd.read_csv(DATA_DIR / DATASET / "results" / "dynamo_KO_single.csv", index_col=0)
pval_KO_dyn = pd.read_csv(DATA_DIR / DATASET / "results" / "dynamo_KO_single_pval.csv", index_col=0)
coef_KO_dyn = coef_KO_dyn.loc[single_ko, TERMINAL_STATES_KO]
pval_KO_dyn = pval_KO_dyn.loc[single_ko, TERMINAL_STATES_KO]
pred_m = coef_KO_dyn - 0.5
pred_m_filter = pred_m[pval_KO_dyn < 0.05]
pred_m_filter[np.isnan(pred_m_filter)] = 0
pred_m_filter.columns = single_matrix.columns
score = np.sum(np.sign(single_matrix) * np.sign(pred_m_filter) == 1).sum() / np.sum(single_matrix != 0).sum()
fate_s.append(score)
method2.append("Dynamo (KO)")
for nrun in range(3):
coef_perturb_rgv = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_single.csv", index_col=0
)
pval_perturb_rgv = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_single_pval.csv", index_col=0
)
coef_perturb_rgv = coef_perturb_rgv.loc[single_ko, TERMINAL_STATES_KO]
pval_perturb_rgv = pval_perturb_rgv.loc[single_ko, TERMINAL_STATES_KO]
pred_m = coef_perturb_rgv
pred_m_filter = pred_m[pval_perturb_rgv < 0.05]
pred_m_filter[np.isnan(pred_m_filter)] = 0
pred_m_filter.columns = single_matrix.columns
score = np.sum(np.sign(single_matrix) * np.sign(pred_m_filter) == 1).sum() / np.sum(single_matrix != 0).sum()
fate_s.append(score)
method2.append("RegVelo")
for nrun in range(3):
coef_perturb_rgv = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_single.csv", index_col=0
)
pval_perturb_rgv = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_single_pval.csv", index_col=0
)
coef_perturb_rgv = coef_perturb_rgv.loc[single_ko, TERMINAL_STATES_KO]
pval_perturb_rgv = pval_perturb_rgv.loc[single_ko, TERMINAL_STATES_KO]
coef_perturb_rgv = coef_perturb_rgv.sample(frac=1).reset_index(drop=True)
pval_perturb_rgv = pval_perturb_rgv.sample(frac=1).reset_index(drop=True)
pred_m = coef_perturb_rgv
pred_m_filter = pred_m[pval_perturb_rgv < 0.05]
pred_m_filter[np.isnan(pred_m_filter)] = 0
pred_m_filter.columns = single_matrix.columns
pred_m_filter.index = single_matrix.index
# shuffled = np.random.permutation(pred_m_filter)
score = np.sum(np.sign(single_matrix) * np.sign(pred_m_filter) == 1).sum() / np.sum(single_matrix != 0).sum()
fate_s.append(score)
method2.append("Random baseline")
Multiple knock-out panel prediction results#
single_matrix = perturbation.loc[single_ko + multiple_ko, :]
pval = pd.read_csv(DATA_DIR / DATASET / "results" / "pval_mat.csv", index_col=0)
pval = pval.loc[single_matrix.index.tolist(), single_matrix.columns.tolist()]
mask_pos = (pval <= 0.05).astype(int)
mask_neg = (pval >= 0.95).astype(int)
single_matrix = mask_neg * (-1) + mask_pos
coef_KO_dyn = pd.read_csv(DATA_DIR / DATASET / "results" / "dynamo_KO_single.csv", index_col=0)
coef_KO_dyn2 = pd.read_csv(DATA_DIR / DATASET / "results" / "dynamo_KO_multiple.csv", index_col=0)
coef_KO_dyn = pd.concat([coef_KO_dyn, coef_KO_dyn2])
coef_KO_dyn = coef_KO_dyn.loc[single_ko + multiple_ko, TERMINAL_STATES_KO]
fate_m = []
method2 = []
pval_KO_dyn = pd.read_csv(DATA_DIR / DATASET / "results" / "dynamo_KO_single_pval.csv", index_col=0)
pval_KO_dyn2 = pd.read_csv(DATA_DIR / DATASET / "results" / "dynamo_KO_multiple_pval.csv", index_col=0)
pval_KO_dyn = pd.concat([pval_KO_dyn, pval_KO_dyn2])
pval_KO_dyn = pval_KO_dyn.loc[single_ko + multiple_ko, TERMINAL_STATES_KO]
pred_m = coef_KO_dyn - 0.5
pred_m_filter = pred_m[pval_KO_dyn <= 0.1]
pred_m_filter[np.isnan(pred_m_filter)] = 0
pred_m_filter.columns = single_matrix.columns
score = np.sum(np.sign(single_matrix) * np.sign(pred_m_filter) == 1).sum() / np.sum(single_matrix != 0).sum()
fate_m.append(score)
method2.append("Dynamo (KO)")
for nrun in range(3):
coef_perturb_rgv = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_single.csv", index_col=0
)
coef_perturb_rgv2 = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_multiple.csv", index_col=0
)
coef_perturb_rgv = pd.concat([coef_perturb_rgv, coef_perturb_rgv2])
pval_perturb_rgv = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_single_pval.csv", index_col=0
)
pval_perturb_rgv2 = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_multiple_pval.csv", index_col=0
)
pval_perturb_rgv = pd.concat([pval_perturb_rgv, pval_perturb_rgv2])
coef_perturb_rgv = coef_perturb_rgv.loc[single_ko + multiple_ko, TERMINAL_STATES_KO]
pval_perturb_rgv = pval_perturb_rgv.loc[single_ko + multiple_ko, TERMINAL_STATES_KO]
pred_m = coef_perturb_rgv
pred_m_filter = pred_m[pval_perturb_rgv <= 0.1]
pred_m_filter[np.isnan(pred_m_filter)] = 0
pred_m_filter.columns = single_matrix.columns
score = np.sum(np.sign(single_matrix) * np.sign(pred_m_filter) == 1).sum() / np.sum(single_matrix != 0).sum()
fate_m.append(score)
method2.append("RegVelo")
for nrun in range(3):
coef_perturb_rgv = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_single.csv", index_col=0
)
coef_perturb_rgv2 = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_multiple.csv", index_col=0
)
coef_perturb_rgv = pd.concat([coef_perturb_rgv, coef_perturb_rgv2])
pval_perturb_rgv = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_single_pval.csv", index_col=0
)
pval_perturb_rgv2 = pd.read_csv(
DATA_DIR / DATASET / "results" / ("runs" + str(nrun + 1)) / "regvelo_multiple_pval.csv", index_col=0
)
pval_perturb_rgv = pd.concat([pval_perturb_rgv, pval_perturb_rgv2])
coef_perturb_rgv = coef_perturb_rgv.loc[single_ko + multiple_ko, TERMINAL_STATES_KO]
pval_perturb_rgv = pval_perturb_rgv.loc[single_ko + multiple_ko, TERMINAL_STATES_KO]
coef_perturb_rgv = coef_perturb_rgv.sample(frac=1).reset_index(drop=True)
pval_perturb_rgv = pval_perturb_rgv.sample(frac=1).reset_index(drop=True)
pred_m = coef_perturb_rgv
pred_m_filter = pred_m[pval_perturb_rgv <= 0.1]
pred_m_filter[np.isnan(pred_m_filter)] = 0
pred_m_filter.columns = single_matrix.columns
pred_m_filter.index = single_matrix.index
# shuffled = np.random.permutation(pred_m_filter)
score = np.sum(np.sign(single_matrix) * np.sign(pred_m_filter) == 1).sum() / np.sum(single_matrix != 0).sum()
fate_m.append(score)
method2.append("Random baseline")
Visualize prediction results#
dat = pd.DataFrame({"cell fate prediction correctness": fate_s, "method": method2})
dat2 = pd.DataFrame({"cell fate prediction correctness": fate_m, "method": method2})
dat["Experimental class"] = "Single TF knock-out"
dat2["Experimental class"] = "Single + Multiple TF knock-out"
df = pd.concat([dat, dat2], axis=0)
METHOD_PALETTE_PERTURBATION["Random baseline"] = "#808080"
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(4, 4))
# Plot the barplot without error bars
sns.barplot(
data=df,
y="Experimental class",
x="cell fate prediction correctness",
hue="method",
hue_order=["RegVelo", "Dynamo (KO)", "Random baseline"],
palette=METHOD_PALETTE_PERTURBATION,
ax=ax,
ci=None,
)
# Add jittered dots
sns.stripplot(
data=df,
y="Experimental class",
x="cell fate prediction correctness",
hue="method",
hue_order=["RegVelo", "Dynamo(KO)", "Random baseline"],
dodge=True,
color="black",
ax=ax,
jitter=True,
)
# Remove the duplicate legend
handles, labels = ax.get_legend_handles_labels()
ax.legend(
handles[4 : (4 + len(METHOD_PALETTE_PERTURBATION))],
labels[4 : (4 + len(METHOD_PALETTE_PERTURBATION))],
bbox_to_anchor=(0.5, -0.1),
loc="upper center",
ncol=2,
)
# Customize labels and other settings
ax.set(ylabel="", xlabel="Cell fate prediction recall")
ax.set_xlabel(xlabel="Cell fate prediction recall", fontsize=13)
# ax.set_xlim(0.45,)
if SAVE_FIGURES:
plt.savefig(
DATA_DIR / DATASET / "results" / "cell_fate_recall.svg", format="svg", transparent=True, bbox_inches="tight"
)
plt.show()