Preprocess mHSPC datasets#
Library imports#
import scanpy as sc
import scvelo as scv
from velovi import preprocess_data
from rgv_tools import DATA_DIR
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_csv from `anndata` is deprecated. Import anndata.io.read_csv instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_excel from `anndata` is deprecated. Import anndata.io.read_excel instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_hdf from `anndata` is deprecated. Import anndata.io.read_hdf instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_loom from `anndata` is deprecated. Import anndata.io.read_loom instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_mtx from `anndata` is deprecated. Import anndata.io.read_mtx instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_text from `anndata` is deprecated. Import anndata.io.read_text instead.
warnings.warn(msg, FutureWarning)
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/anndata/utils.py:429: FutureWarning: Importing read_umi_tools from `anndata` is deprecated. Import anndata.io.read_umi_tools instead.
warnings.warn(msg, FutureWarning)
General settings#
sc.settings.verbosity = 2
scv.settings.verbosity = 3
Constants#
DATASET = "mHSPC"
SAVE_DATA = True
if SAVE_DATA:
(DATA_DIR / DATASET / "processed").mkdir(parents=True, exist_ok=True)
Data loading#
adata_velo = sc.read_loom(DATA_DIR / DATASET / "raw" / "Nestorowa.loom")
adata_p = sc.read_h5ad(DATA_DIR / DATASET / "raw" / "adata_nestorowa.h5ad")
adata_p.obs
| cell_IDs | cell_types_IDs | cell_types_broad | cell_types_fine | cell_types_broad_cleaned | n_counts | n_genes | mito_ratio | initial_size_spliced | initial_size_unspliced | initial_size | size_factors_x | size_factors_s | size_factors_u | leiden | velocity_self_transition | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SRR3558184 | HSPC_001_rep | HSPC | MPP_broad, STHSC_broad | None | MPP_broad | 1404060.0 | 5826 | 0.0 | 1404060 | 224383 | 1404060.0 | 2.722215 | 2.722215 | 2.770269 | 1 | 0.113736 |
| SRR3558185 | HSPC_002_rep | HSPC | LTHSC_broad | LTHSC | LTHSC_broad | 130334.0 | 4730 | 0.0 | 130334 | 19788 | 130334.0 | 0.242590 | 0.242590 | 0.211278 | 1 | 0.106622 |
| SRR3558186 | HSPC_003_rep | HSPC | MPP_broad, MPP3_broad | None | MPP_broad | 627148.0 | 6372 | 0.0 | 627148 | 84188 | 627148.0 | 1.043037 | 1.043037 | 1.068530 | 3 | 0.022092 |
| SRR3558187 | HSPC_004_rep | HSPC | LMPP_broad | LMPP | LMPP_broad | 317301.0 | 5536 | 0.0 | 317301 | 54361 | 317301.0 | 0.548139 | 0.548139 | 0.602953 | 0 | 0.158245 |
| SRR3558189 | HSPC_006_rep | HSPC | LMPP_broad | LMPP | LMPP_broad | 335563.0 | 6270 | 0.0 | 335563 | 62426 | 335563.0 | 0.612579 | 0.612579 | 0.806794 | 0 | 0.147797 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| SRR3560099 | Prog_848_rep | Prog | GMP_broad | GMP | GMP_broad | 194009.0 | 5750 | 0.0 | 194009 | 39021 | 194009.0 | 0.340196 | 0.340196 | 0.453426 | 9 | 0.174135 |
| SRR3560100 | Prog_849_rep | Prog | CMP_broad | CMP | CMP_broad | 240624.0 | 5787 | 0.0 | 240624 | 32157 | 240624.0 | 0.442644 | 0.442644 | 0.396637 | 6 | 0.084609 |
| SRR3560101 | Prog_850_rep | Prog | MEP_broad | None | MEP_broad | 608044.0 | 7619 | 0.0 | 608044 | 41827 | 608044.0 | 0.868808 | 0.868808 | 0.641310 | 2 | 0.010177 |
| SRR3560102 | Prog_851_rep | Prog | CMP_broad | CMP | CMP_broad | 323195.0 | 6473 | 0.0 | 323195 | 86099 | 323195.0 | 0.534781 | 0.534781 | 1.087060 | 6 | 0.129851 |
| SRR3560103 | Prog_852_rep | Prog | MEP_broad | MEP | MEP_broad | 516834.0 | 7249 | 0.0 | 516834 | 35891 | 516834.0 | 0.788027 | 0.788027 | 0.523927 | 4 | 0.212272 |
1772 rows × 16 columns
Preprocessing#
adata_velo.obs_names[:5]
Index(['onefilepercell_SRR3556264Aligned_and_others_KPB9A:SRR3556264Aligned.out.bam',
'onefilepercell_SRR3556264Aligned_and_others_KPB9A:SRR3556265Aligned.out.bam',
'onefilepercell_SRR3556264Aligned_and_others_KPB9A:SRR3556266Aligned.out.bam',
'onefilepercell_SRR3556264Aligned_and_others_KPB9A:SRR3556267Aligned.out.bam',
'onefilepercell_SRR3556264Aligned_and_others_KPB9A:SRR3556268Aligned.out.bam'],
dtype='object', name='CellID')
adata_velo.obs_names = [
i.replace("onefilepercell_SRR3556264Aligned_and_others_KPB9A:", "") for i in adata_velo.obs_names
]
adata_velo.obs_names = [i.replace("Aligned.out.bam", "") for i in adata_velo.obs_names]
adata_velo = adata_velo[adata_p.obs_names].copy()
adata_velo.obs_names = adata_p.obs["cell_IDs"]
adata_velo.obs_names = [i.replace("_rep", "") for i in adata_velo.obs_names]
adata_velo.obs_names
Index(['HSPC_001', 'HSPC_002', 'HSPC_003', 'HSPC_004', 'HSPC_006', 'HSPC_008',
'HSPC_009', 'HSPC_011', 'HSPC_012', 'HSPC_013',
...
'Prog_843', 'Prog_844', 'Prog_845', 'Prog_846', 'Prog_847', 'Prog_848',
'Prog_849', 'Prog_850', 'Prog_851', 'Prog_852'],
dtype='object', length=1772)
adata_velo.var_names_make_unique()
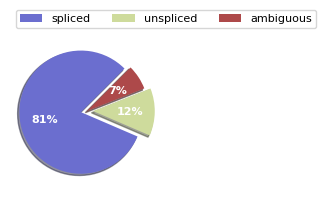
scv.pl.proportions(adata_velo)
scv.pp.filter_and_normalize(adata_velo, min_shared_counts=20, n_top_genes=3000)
Filtered out 18356 genes that are detected 20 counts (shared).
Normalized count data: X, spliced, unspliced.
Extracted 3000 highly variable genes.
Logarithmized X.
/home/icb/weixu.wang/miniconda3/envs/regvelo_test/lib/python3.10/site-packages/scvelo/preprocessing/utils.py:705: DeprecationWarning: `log1p` is deprecated since scVelo v0.3.0 and will be removed in a future version. Please use `log1p` from `scanpy.pp` instead.
log1p(adata)
adata_velo.obs_names = [i.replace("-", "_") for i in adata_velo.obs_names]
Processing Dataset#
adata_subset = adata_velo.copy()
## set annotation
adata_p.obs_names = adata_velo.obs_names
adata_subset.obs = adata_p.obs
print(adata_subset)
## Velocity pipeline
sc.tl.pca(adata_subset)
sc.pp.neighbors(adata_subset, n_neighbors=30, n_pcs=30)
scv.pp.moments(adata_subset)
adata_subset = preprocess_data(adata_subset, filter_on_r2=True)
if SAVE_DATA:
adata_subset.write_h5ad(DATA_DIR / DATASET / "processed" / "mHSC_ExpressionData.h5ad")
AnnData object with n_obs × n_vars = 1772 × 3000
obs: 'cell_IDs', 'cell_types_IDs', 'cell_types_broad', 'cell_types_fine', 'cell_types_broad_cleaned', 'n_counts', 'n_genes', 'mito_ratio', 'initial_size_spliced', 'initial_size_unspliced', 'initial_size', 'size_factors_x', 'size_factors_s', 'size_factors_u', 'leiden', 'velocity_self_transition'
var: 'Accession', 'Chromosome', 'End', 'Start', 'Strand', 'gene_count_corr', 'means', 'dispersions', 'dispersions_norm', 'highly_variable'
uns: 'log1p'
layers: 'matrix', 'ambiguous', 'spanning', 'spliced', 'unspliced'
computing PCA
with n_comps=50
finished (0:00:02)
computing neighbors
using 'X_pca' with n_pcs = 30
2025-10-25 11:03:55.896387: E external/local_xla/xla/stream_executor/cuda/cuda_fft.cc:477] Unable to register cuFFT factory: Attempting to register factory for plugin cuFFT when one has already been registered
WARNING: All log messages before absl::InitializeLog() is called are written to STDERR
E0000 00:00:1761383037.019661 1275922 cuda_dnn.cc:8310] Unable to register cuDNN factory: Attempting to register factory for plugin cuDNN when one has already been registered
E0000 00:00:1761383037.440171 1275922 cuda_blas.cc:1418] Unable to register cuBLAS factory: Attempting to register factory for plugin cuBLAS when one has already been registered
finished (0:01:55)
computing moments based on connectivities
finished (0:00:00) --> added
'Ms' and 'Mu', moments of un/spliced abundances (adata.layers)
computing velocities
finished (0:00:00) --> added
'velocity', velocity vectors for each individual cell (adata.layers)